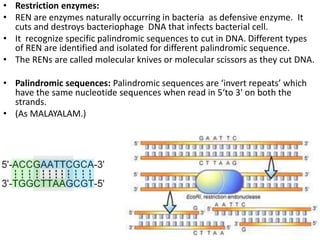
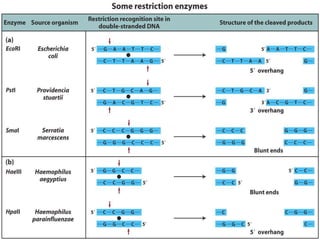
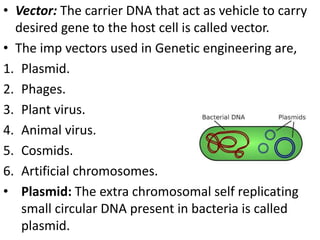
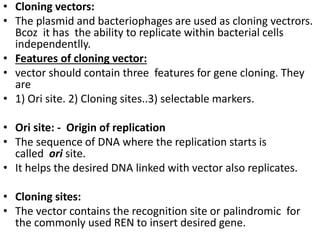
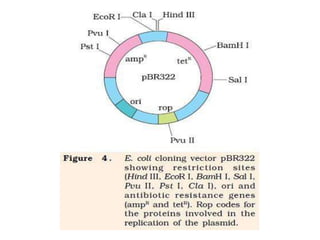
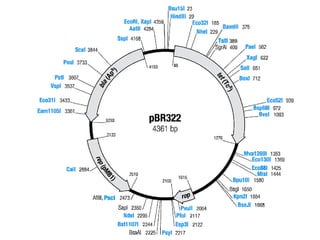
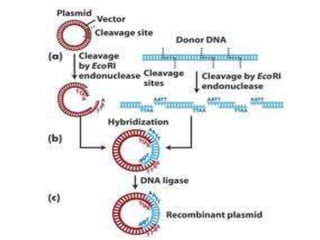
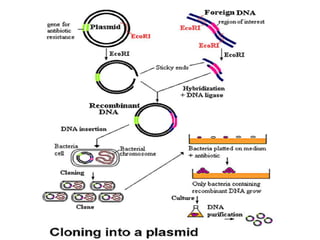
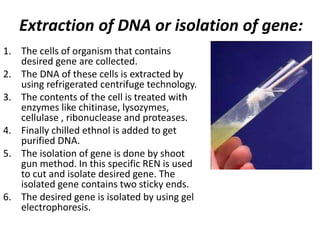
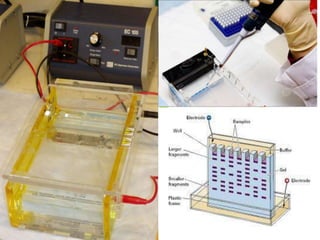
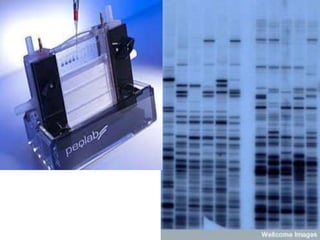
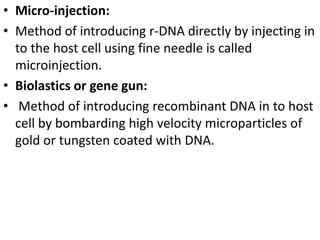
This document discusses biotechnology and genetic engineering techniques. It explains that biotechnology uses organisms or enzymes to produce useful products. Genetic engineering techniques allow modification of genetic material like DNA and RNA to change host organism phenotypes. Key techniques include identifying genes of interest, introducing them into hosts, and maintaining the introduced DNA in progeny. Restriction enzymes and vectors are important tools that allow cutting and recombining of DNA to clone genes and transfer them to target organisms.