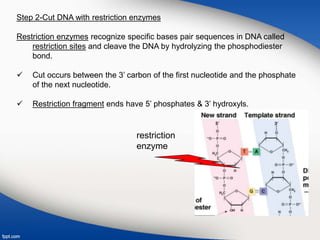
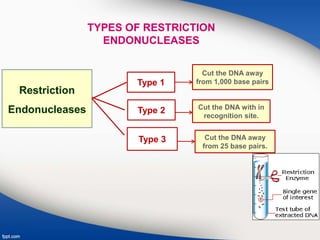
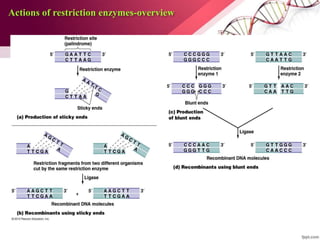
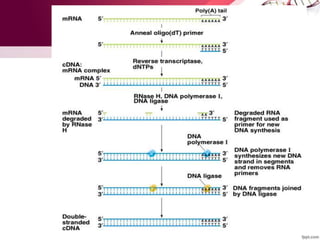
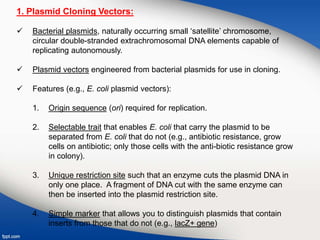
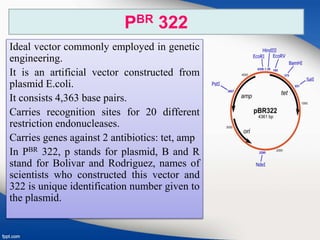
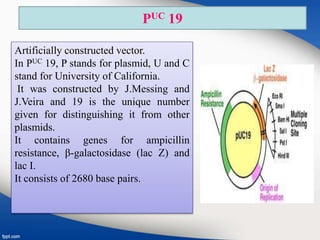
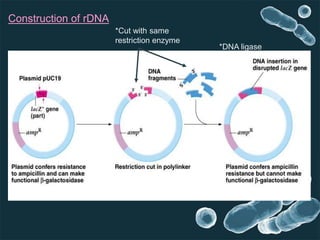
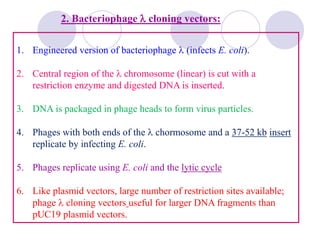
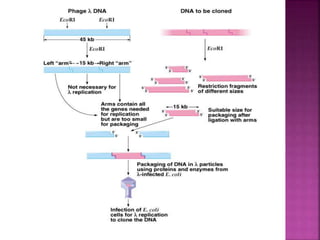
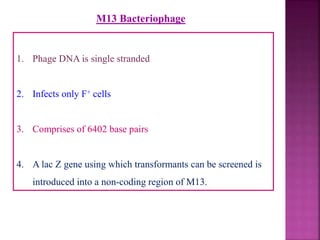
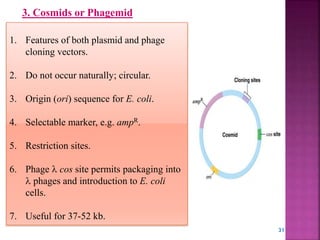
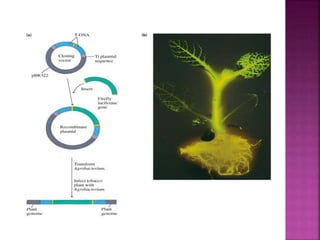
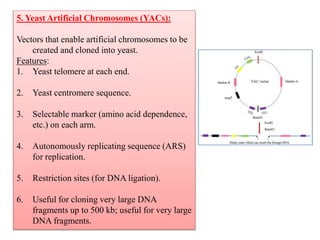
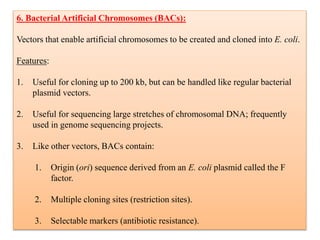
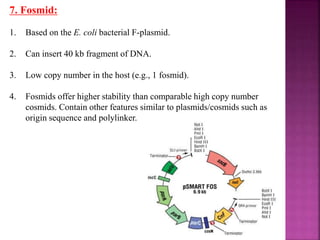
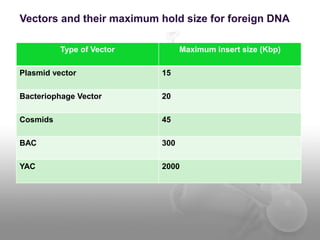
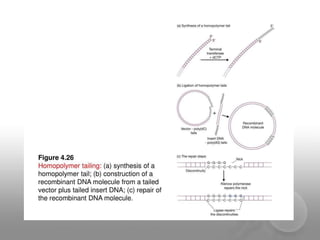
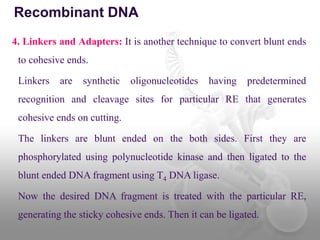
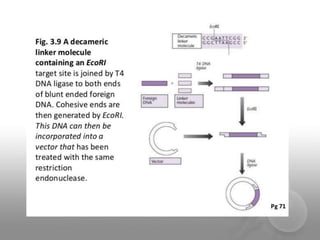
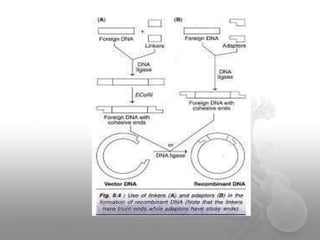
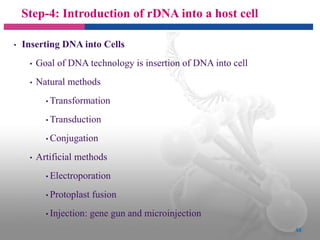
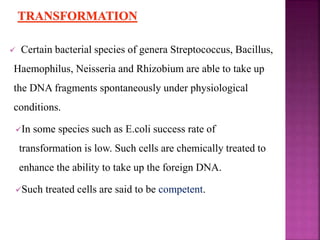
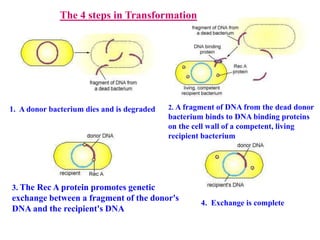
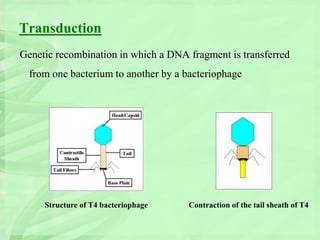
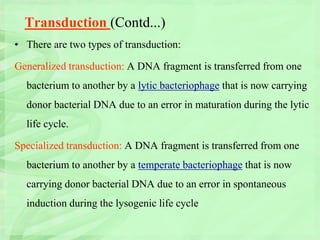
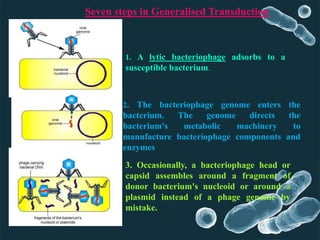
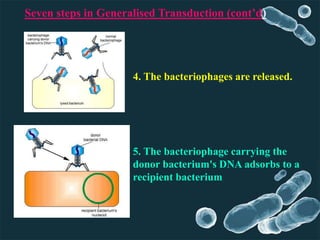
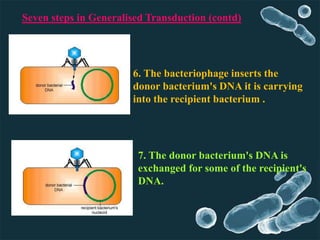
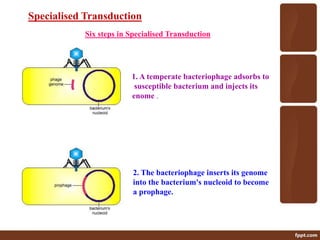
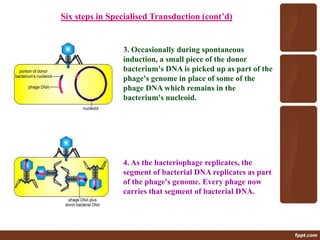
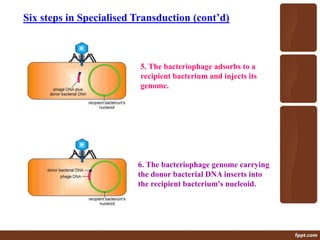
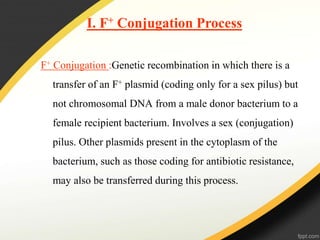
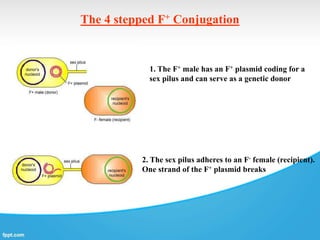
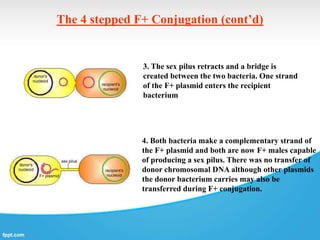
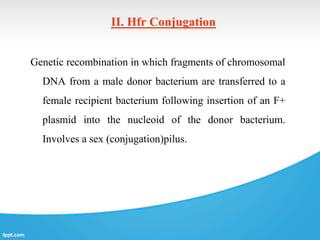
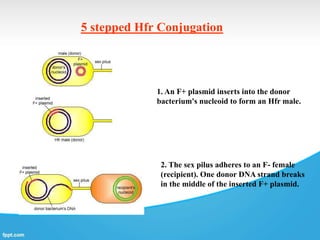
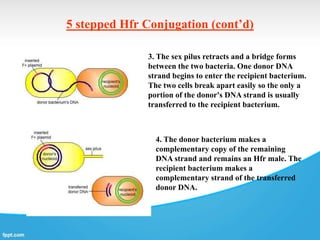
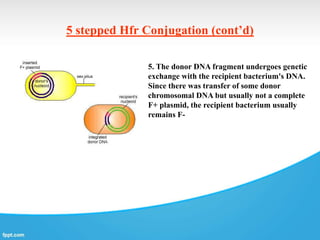
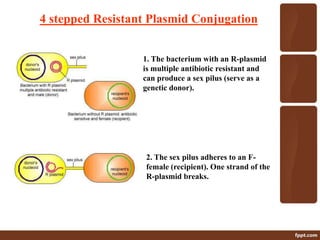
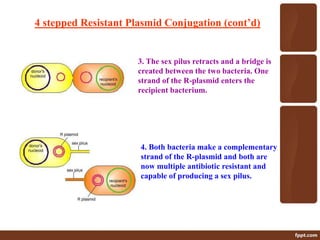
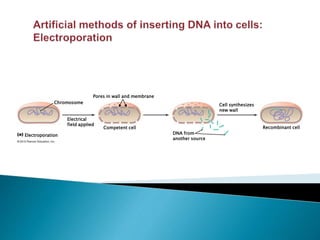
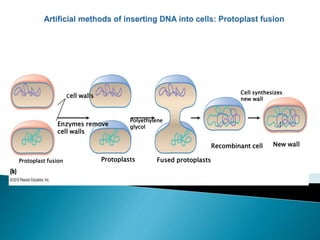
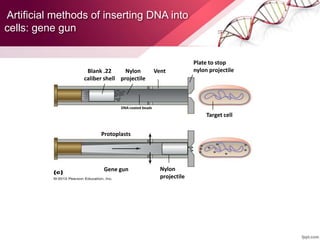
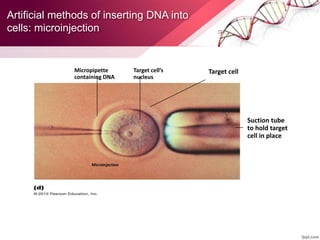
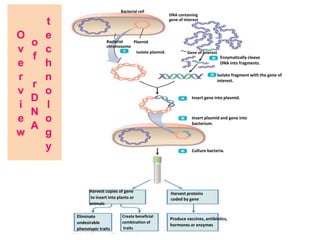
Genetic engineering involves directly manipulating genes, often by adding a gene from another species to an organism's genome. This is done through recombinant DNA (rDNA) technology, which combines DNA sequences artificially. A key part of the process is using restriction enzymes to cut DNA at specific sites, then inserting the cut DNA fragment into a vector like a plasmid for replication in a host cell. The engineered DNA is then introduced into host cells, and cells containing the new DNA are identified and isolated through markers on the vector.