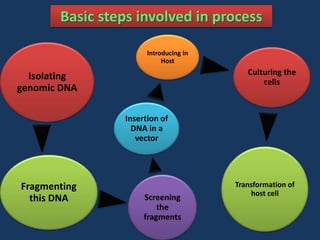
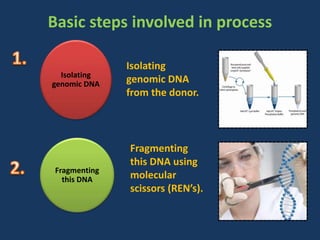
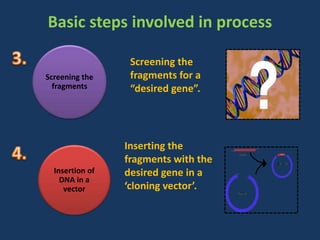
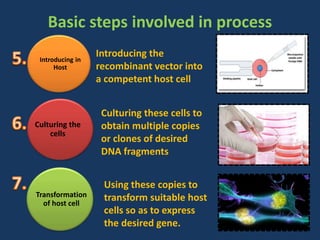
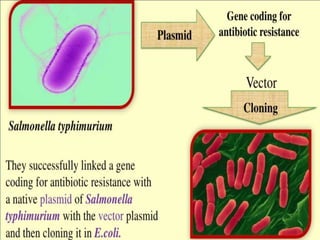
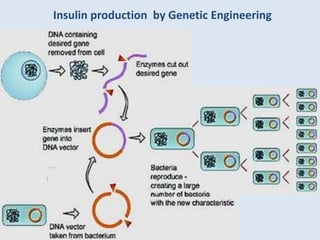
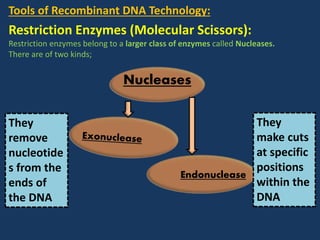
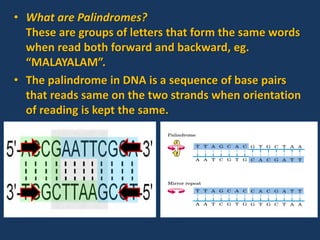
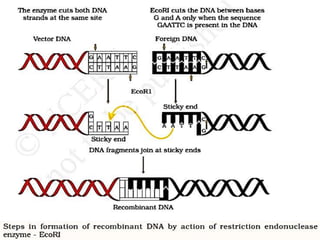
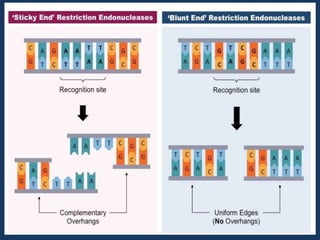
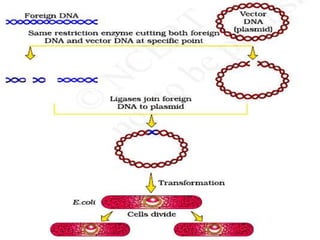
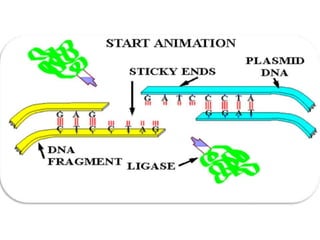
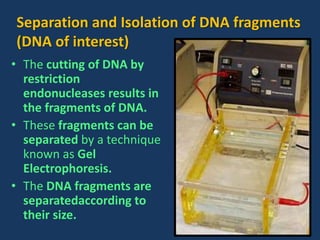
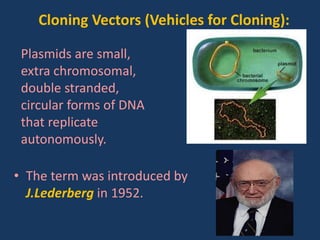
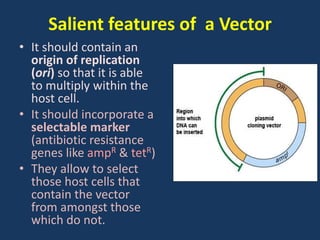
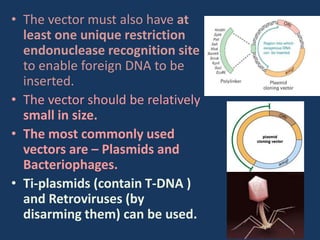
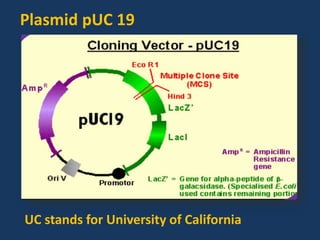
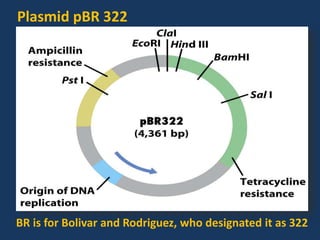
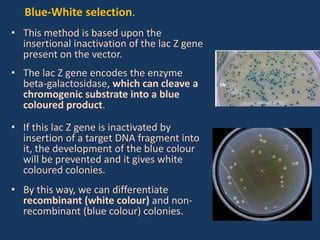
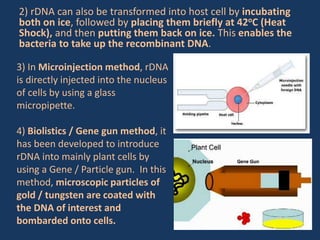
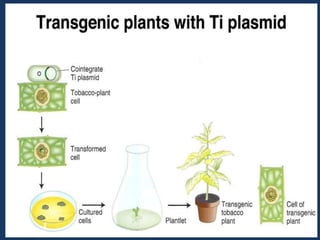
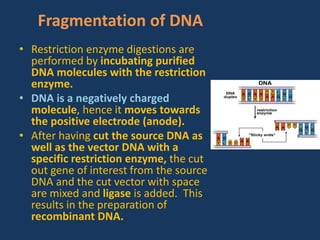
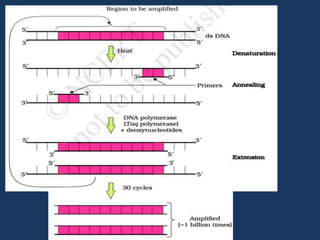
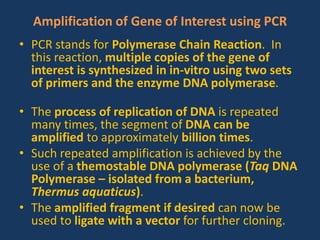
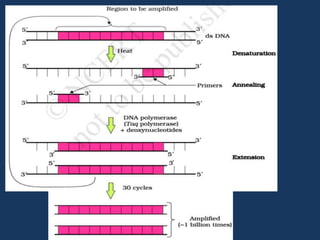
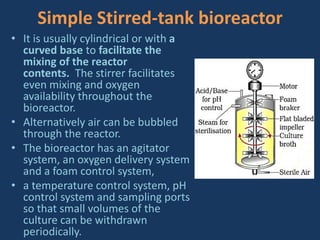
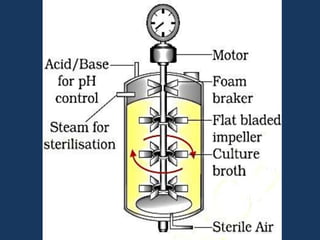
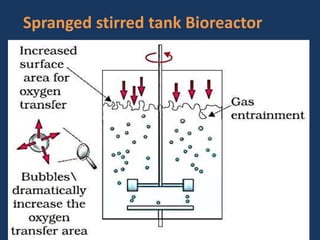
The document discusses biotechnology and recombinant DNA technology. It defines biotechnology as using organisms or enzymes from organisms to produce useful products. Recombinant DNA technology involves isolating DNA, fragmenting it with enzymes, inserting fragments into vectors, transforming host cells, and culturing the cells to multiply the DNA. The basic steps are isolating a gene, inserting it into a vector, introducing the vector into a host cell, and using the host to generate multiple copies of the gene.