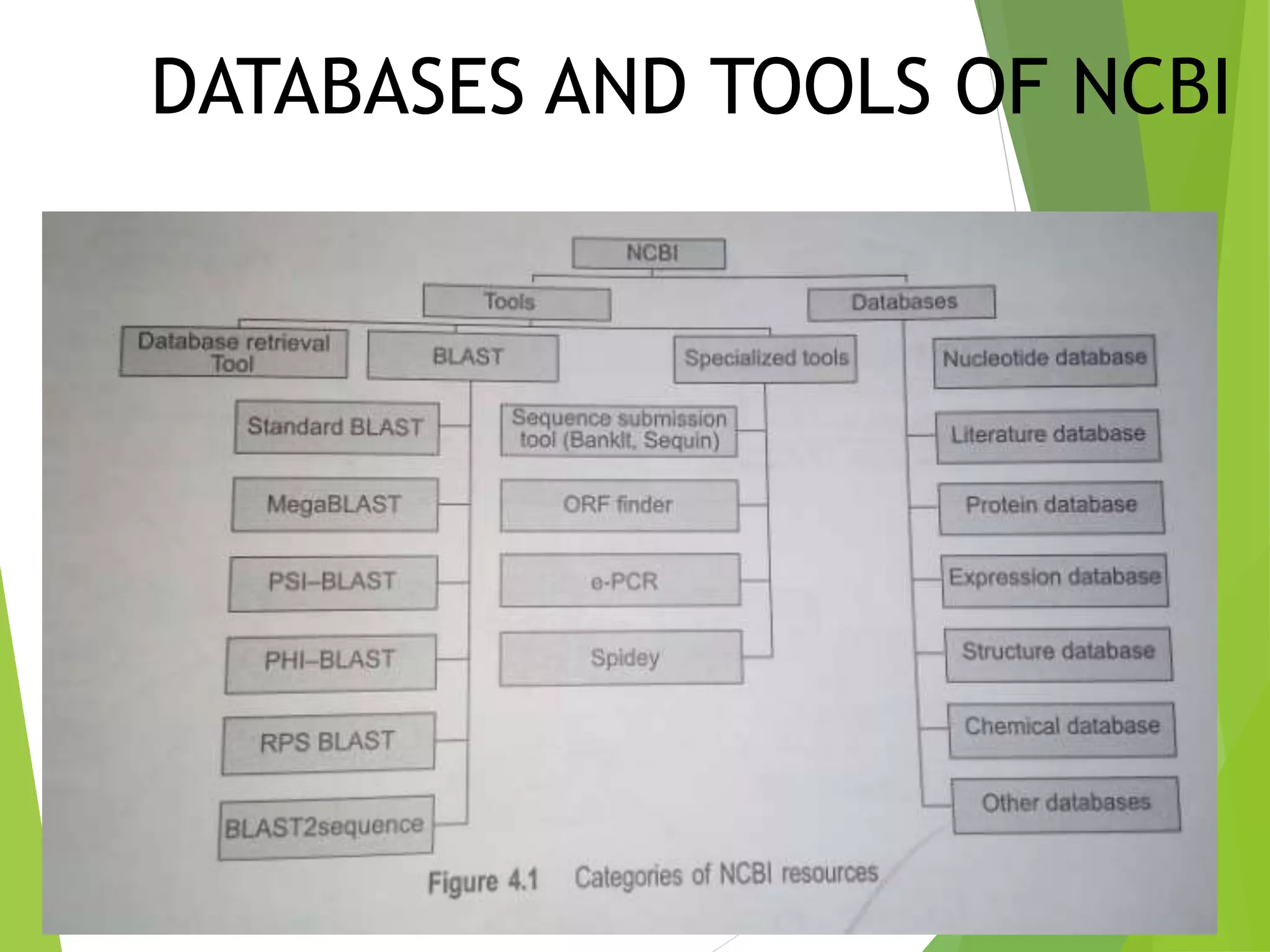
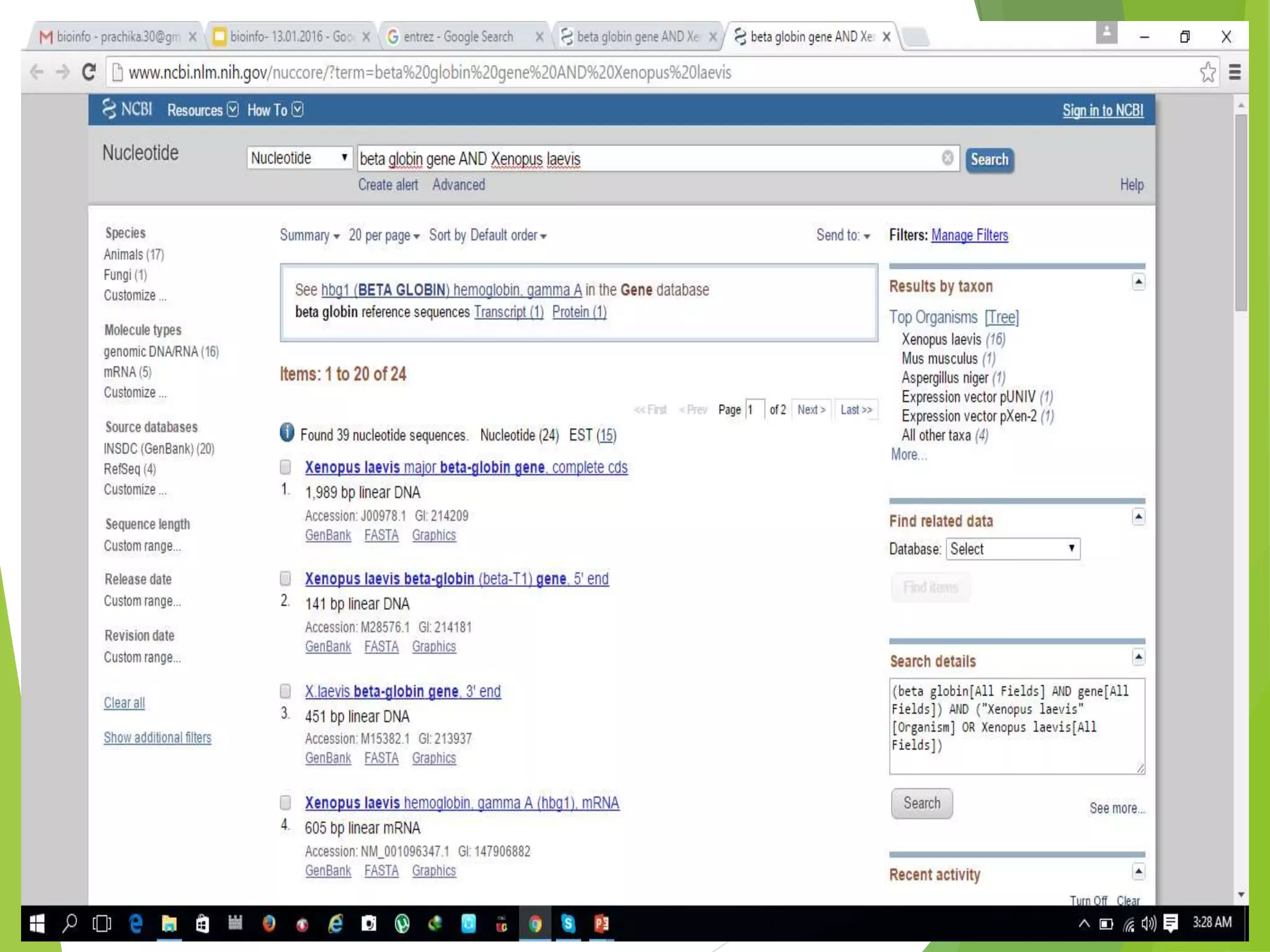
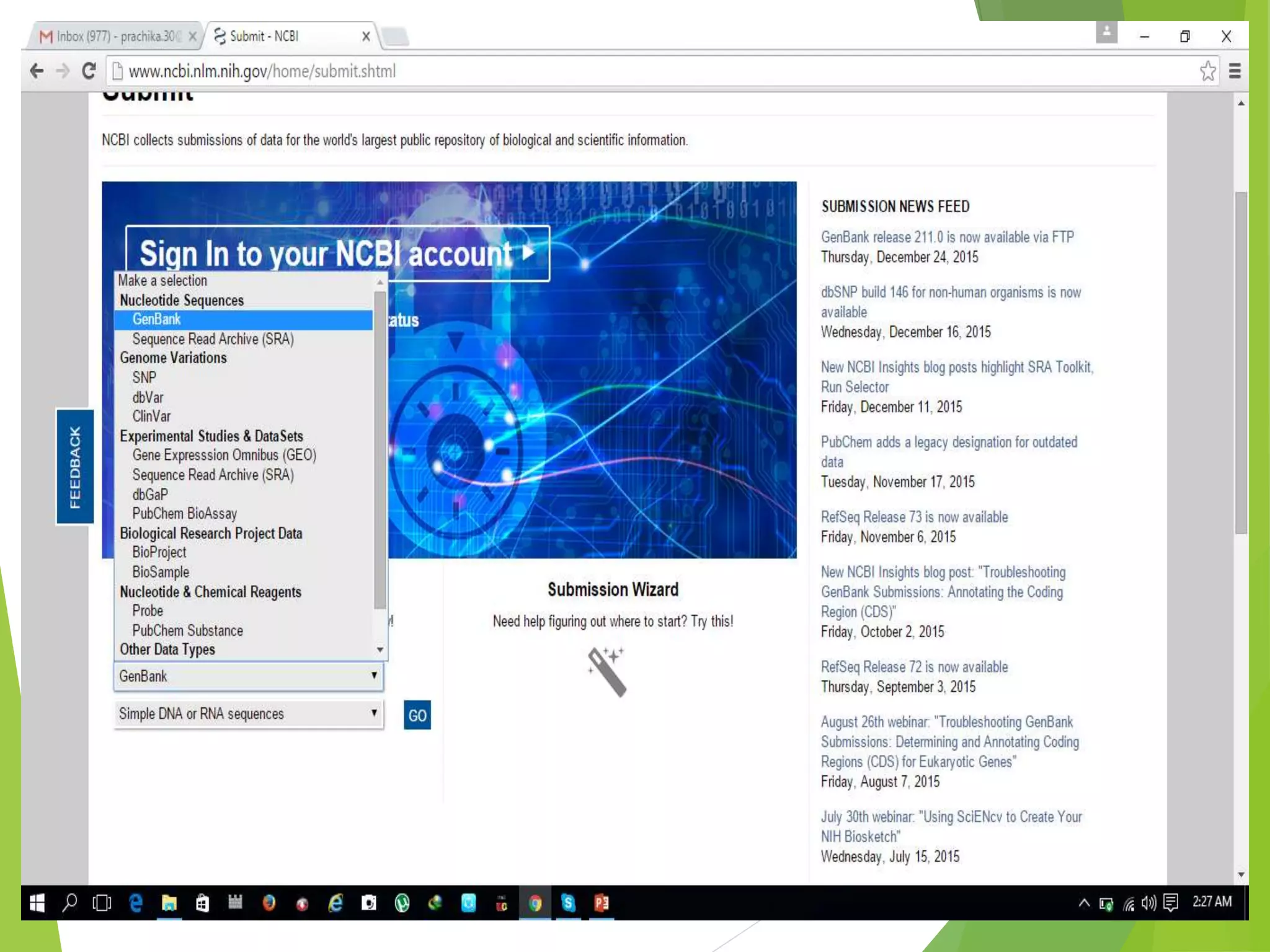
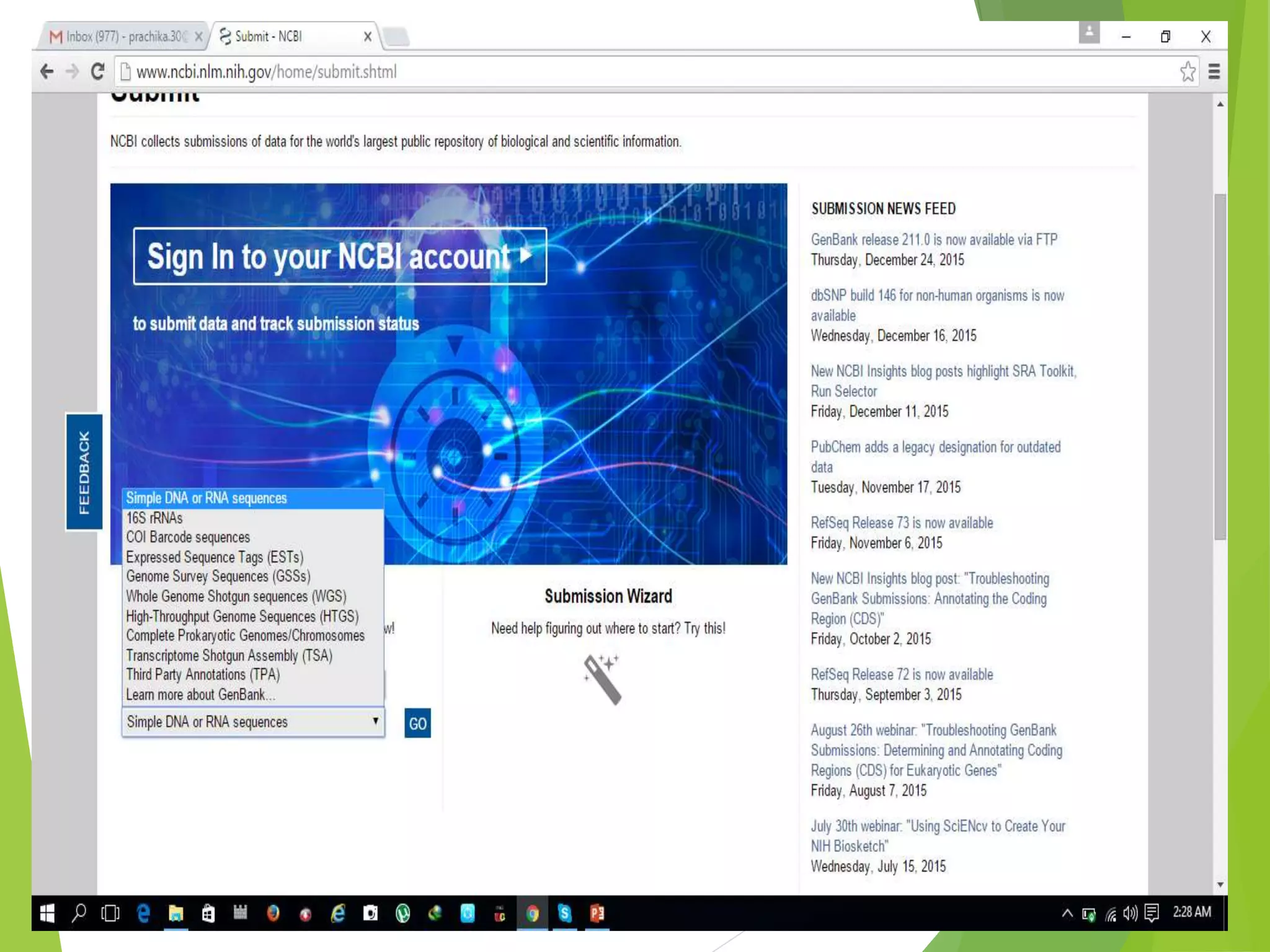
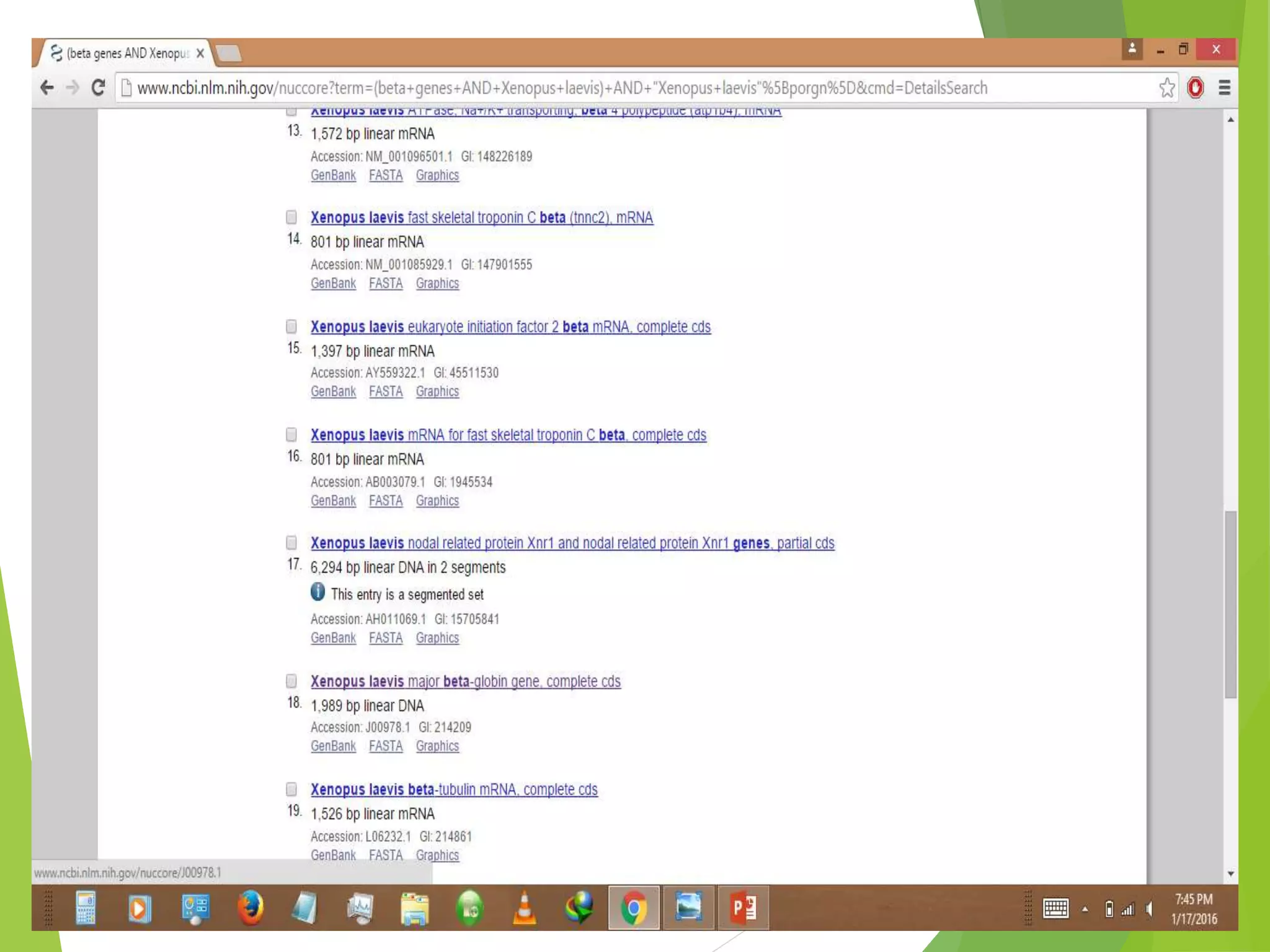
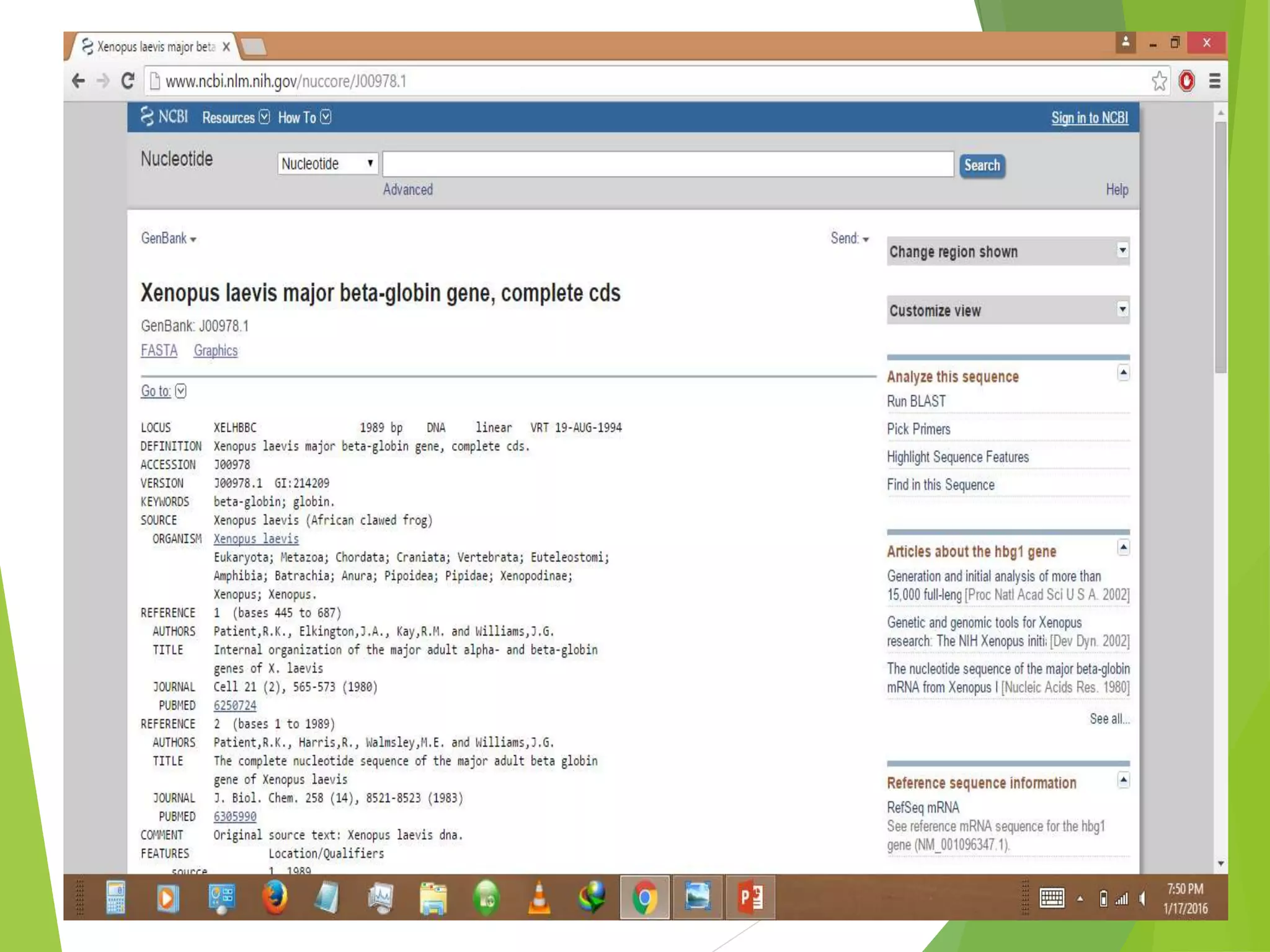
The National Center for Biotechnology Information (NCBI) was established in 1988 as part of the National Library of Medicine. NCBI houses numerous biomedical databases including those related to genes, proteins, molecular structures, gene expression, and biomedical literature. Users can utilize various tools on the NCBI site to search databases, perform sequence alignments using BLAST, and submit new sequences. Some key databases include GenBank (nucleotide sequences), PubMed (biomedical literature), and RefSeq (non-redundant reference sequences).