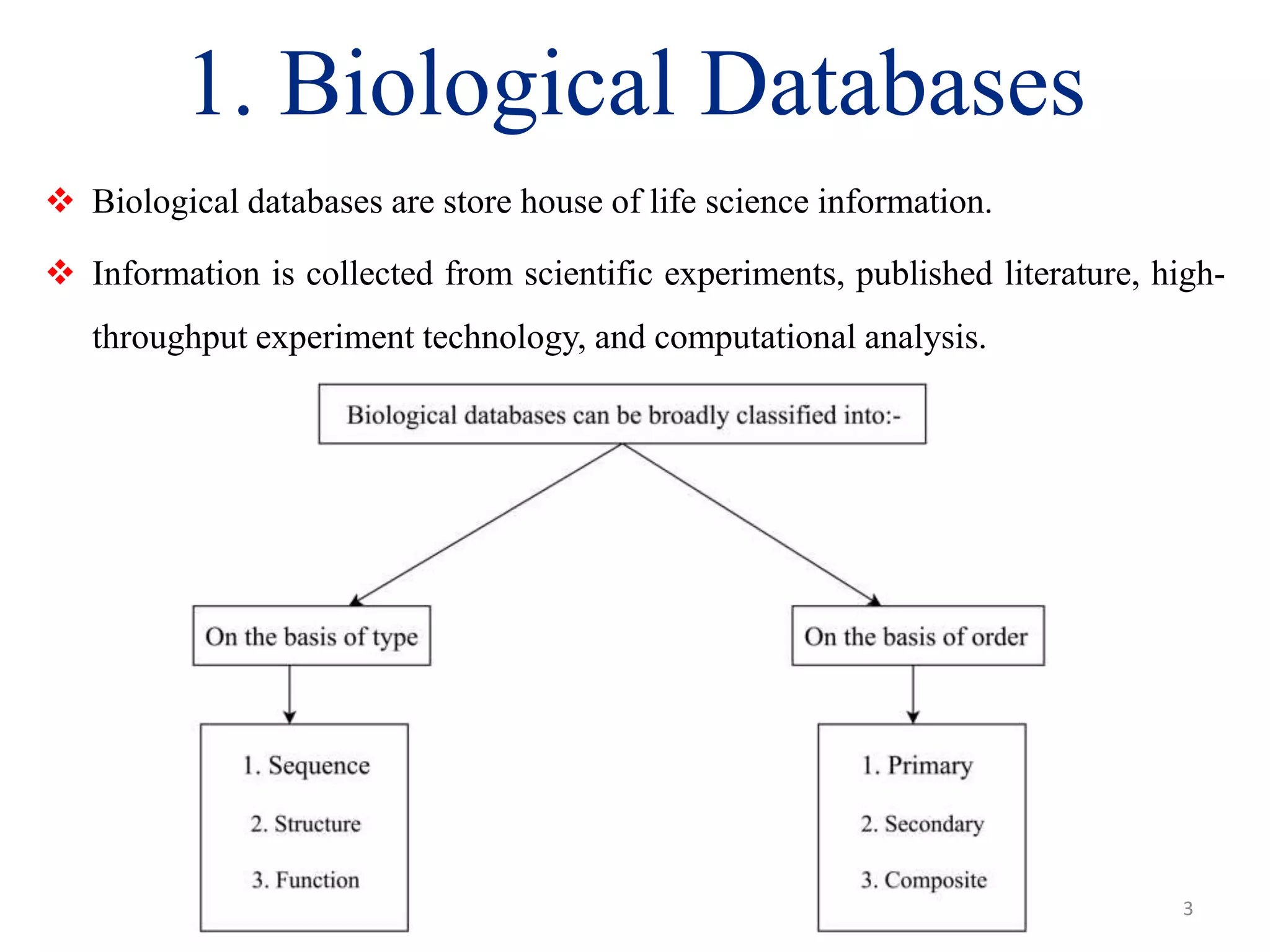
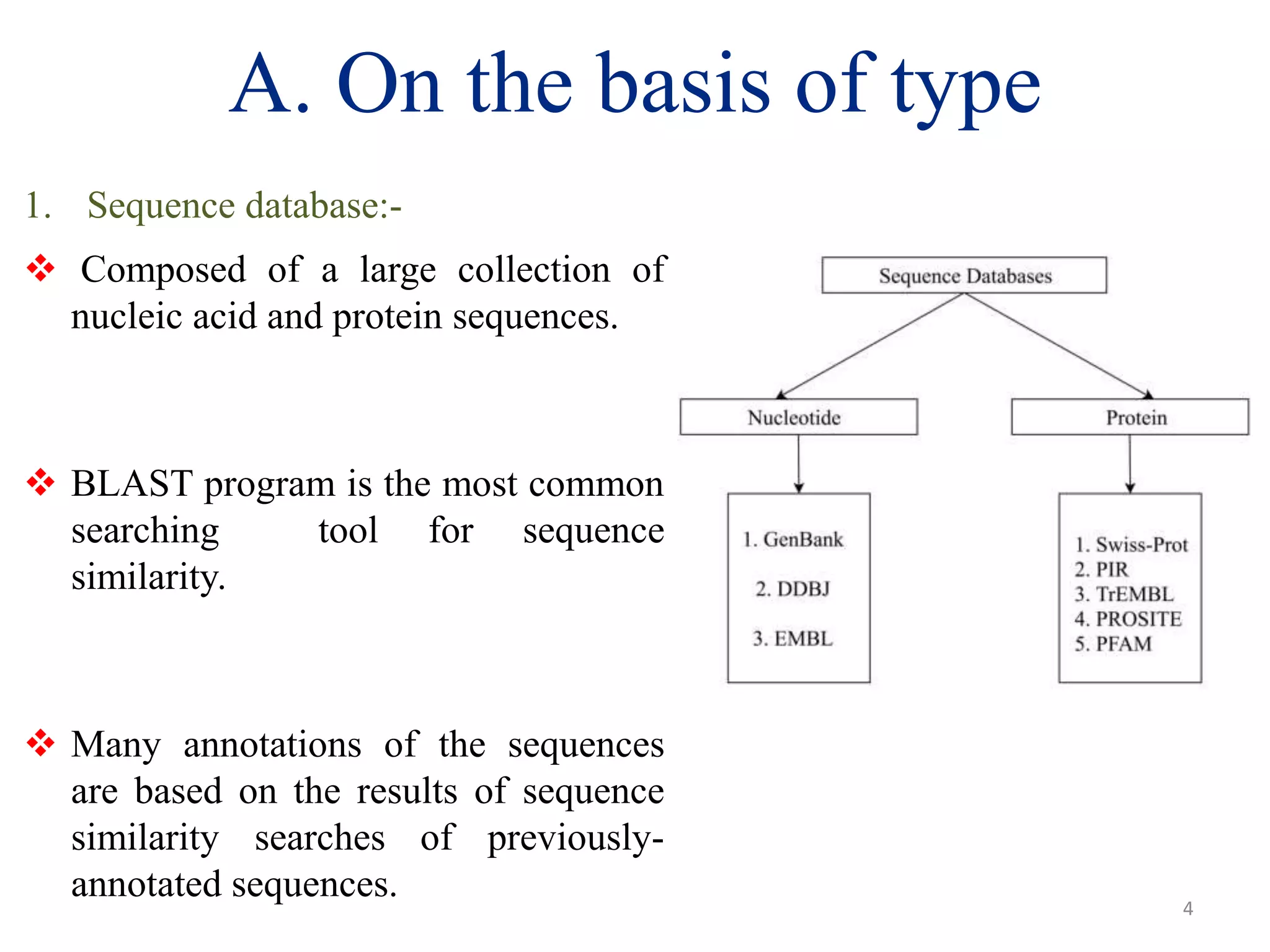
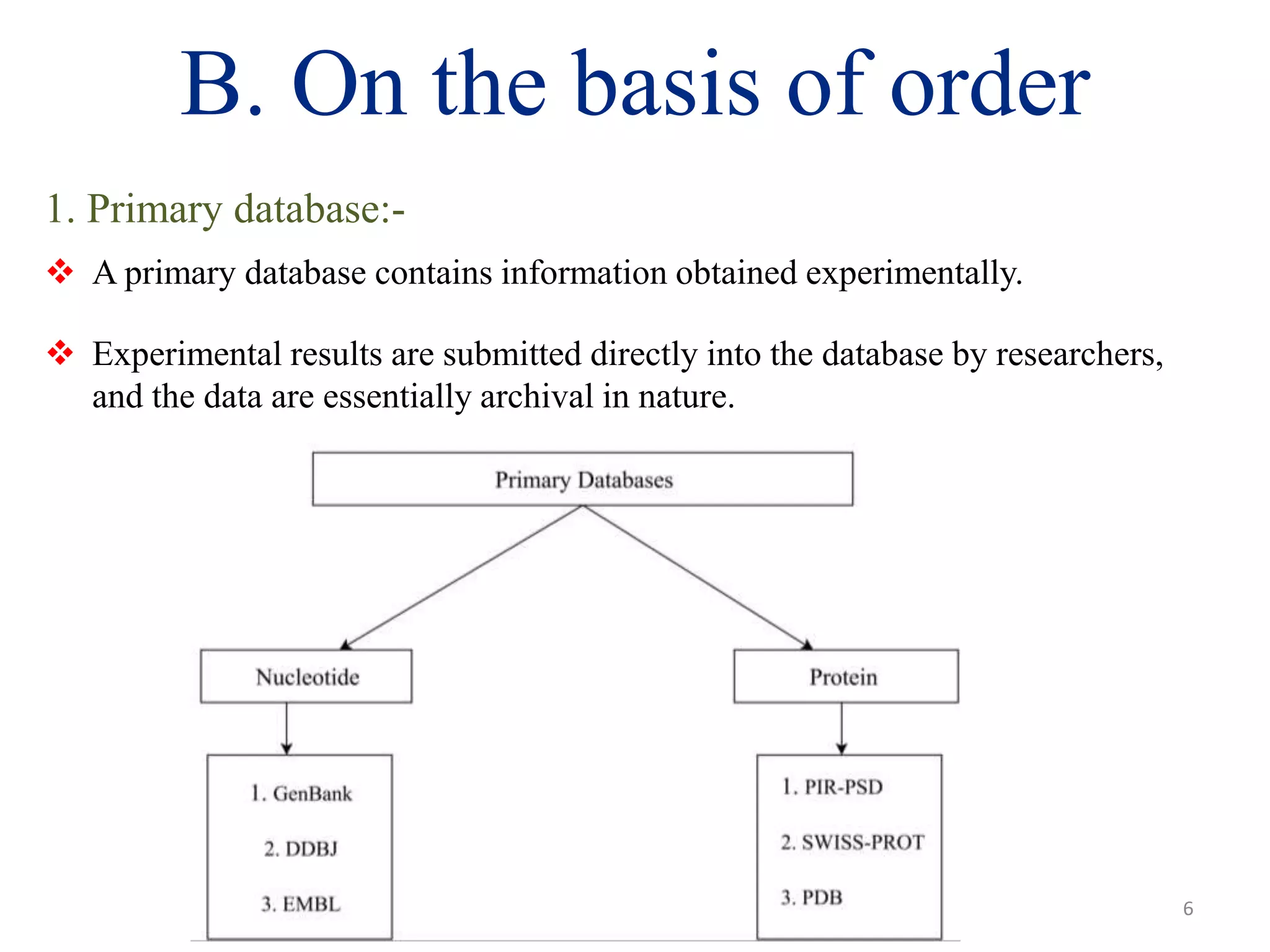
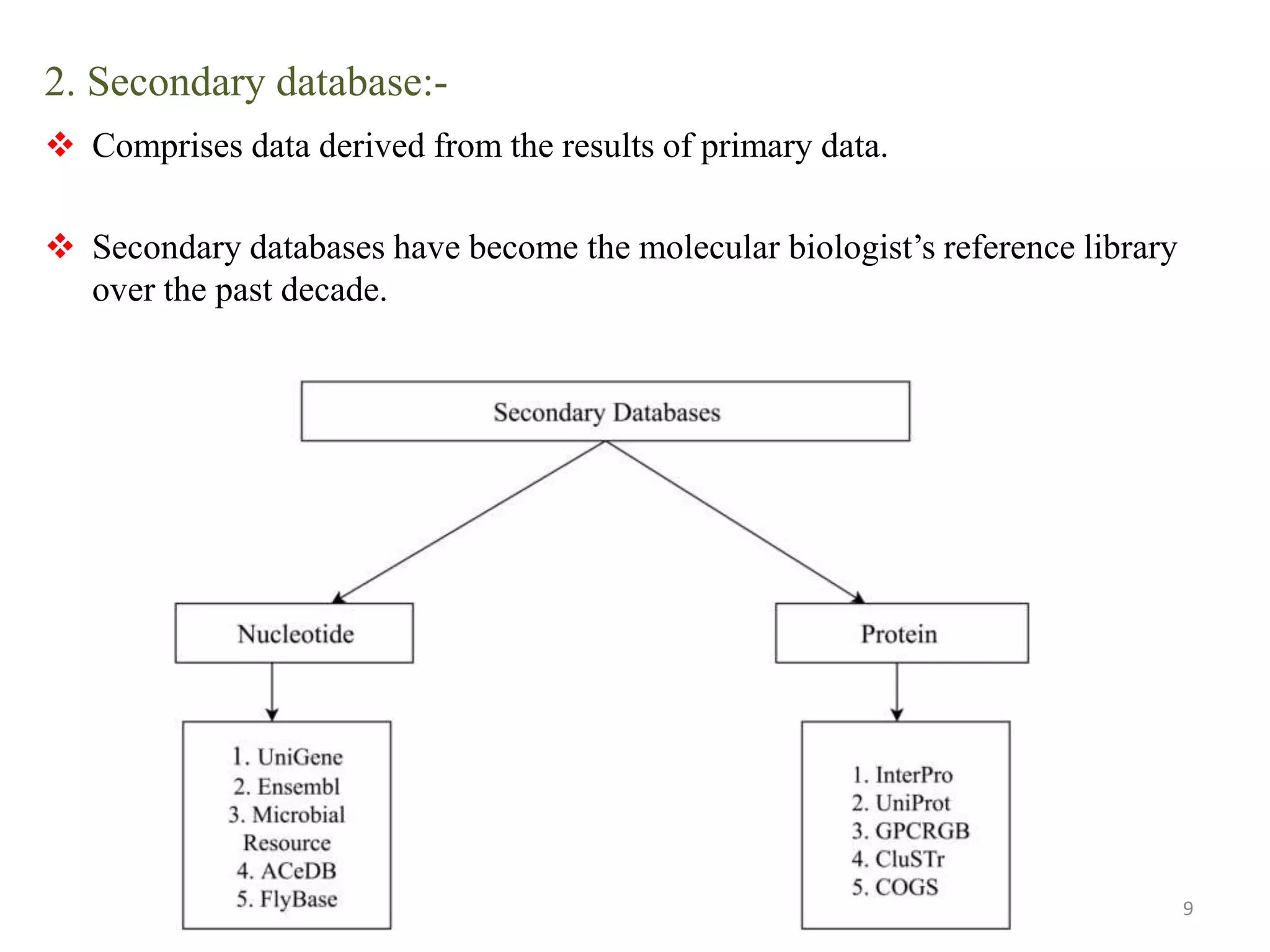
The document discusses biological databases, categorizing them into sequence, structure, and functional databases based on their type, as well as primary, secondary, and composite databases based on order. It provides examples of each type, detailing how they store life science information obtained from experiments and literature. The conclusion highlights the challenges in managing vast data volumes and the importance of bioinformatics in advancing biological sciences.