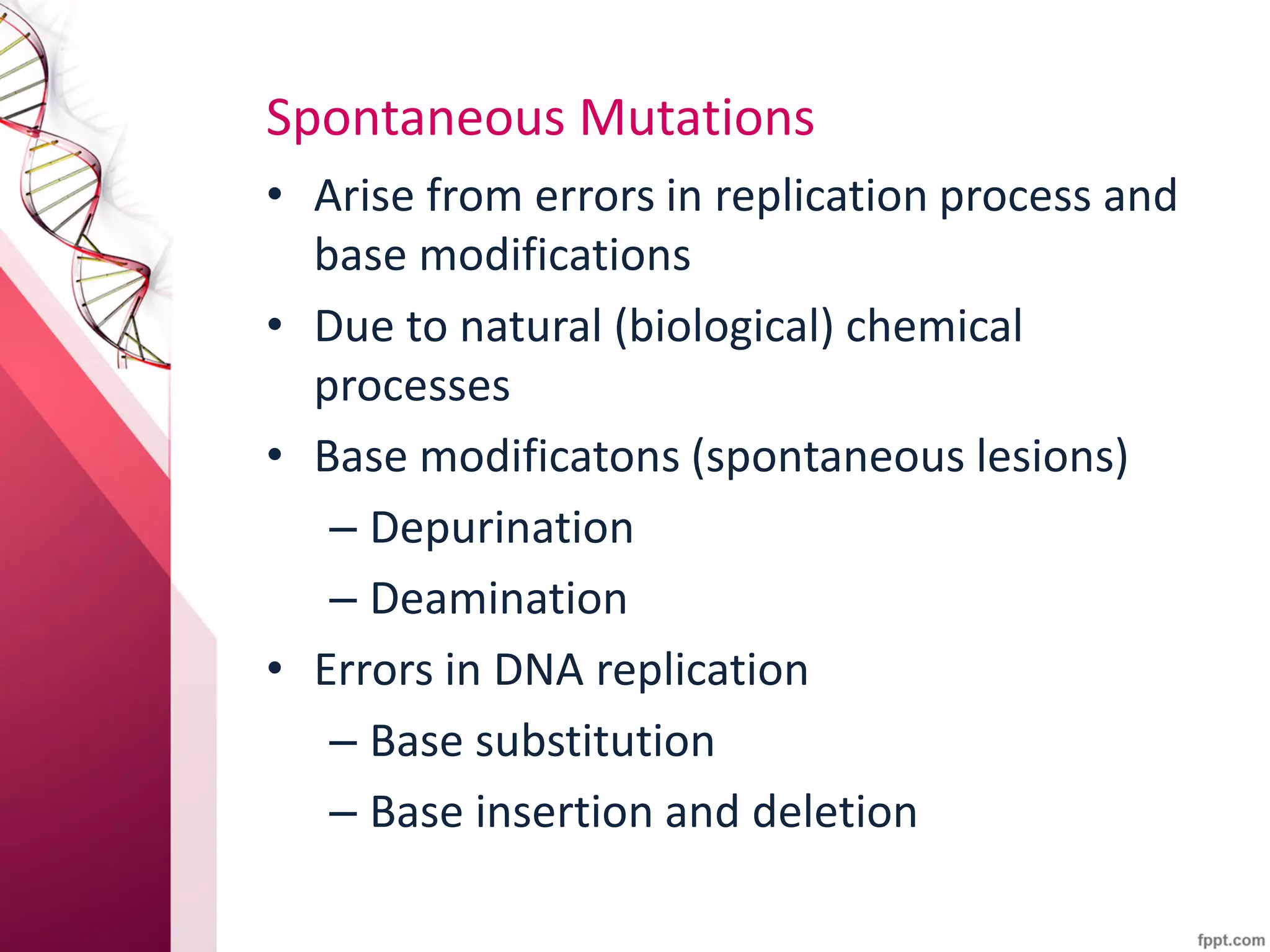
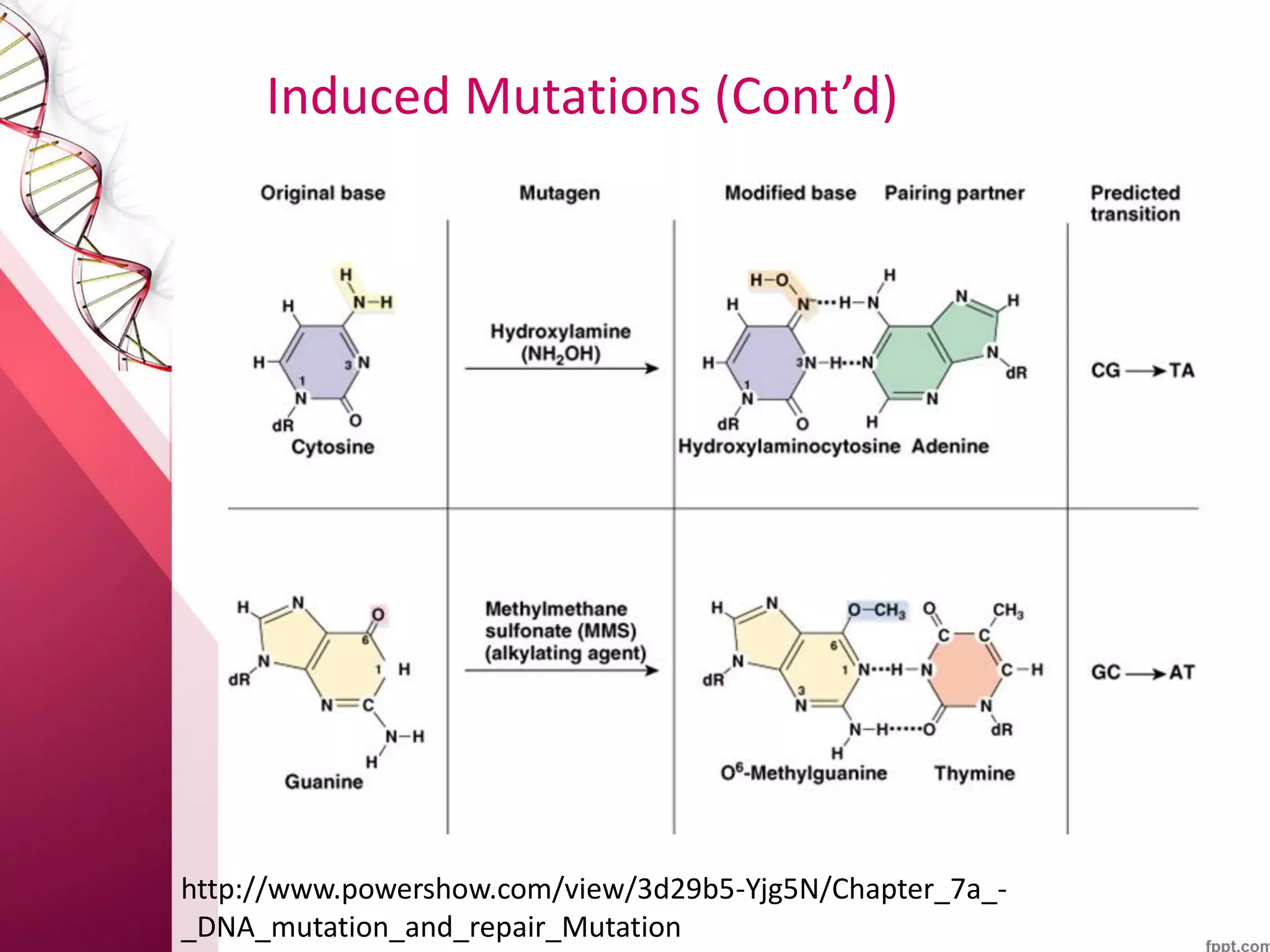
1) DNA mutations can occur spontaneously from errors in replication or due to environmental mutagens, and can lead to genetic disorders if unrepaired.
2) The cell has multiple DNA repair mechanisms to recognize and fix damage, such as base excision repair and nucleotide excision repair. Failure to repair can increase mutations.
3) The study found that DNA damage increased phosphorylation and nuclear localization of the tumor suppressor PTEN, which plays a role in DNA repair and maintaining genomic integrity. Phosphorylation of PTEN was associated with enhanced DNA repair.