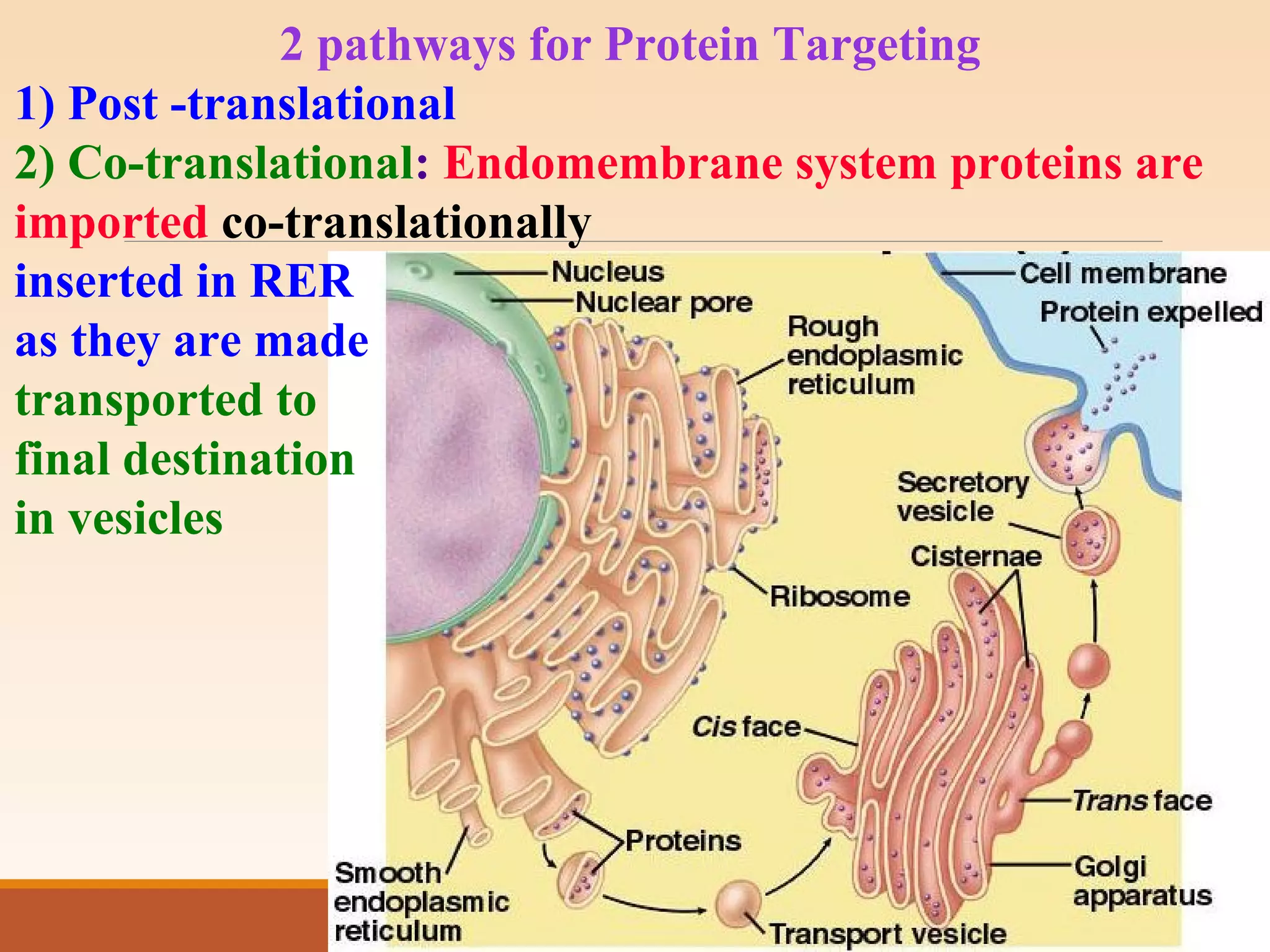
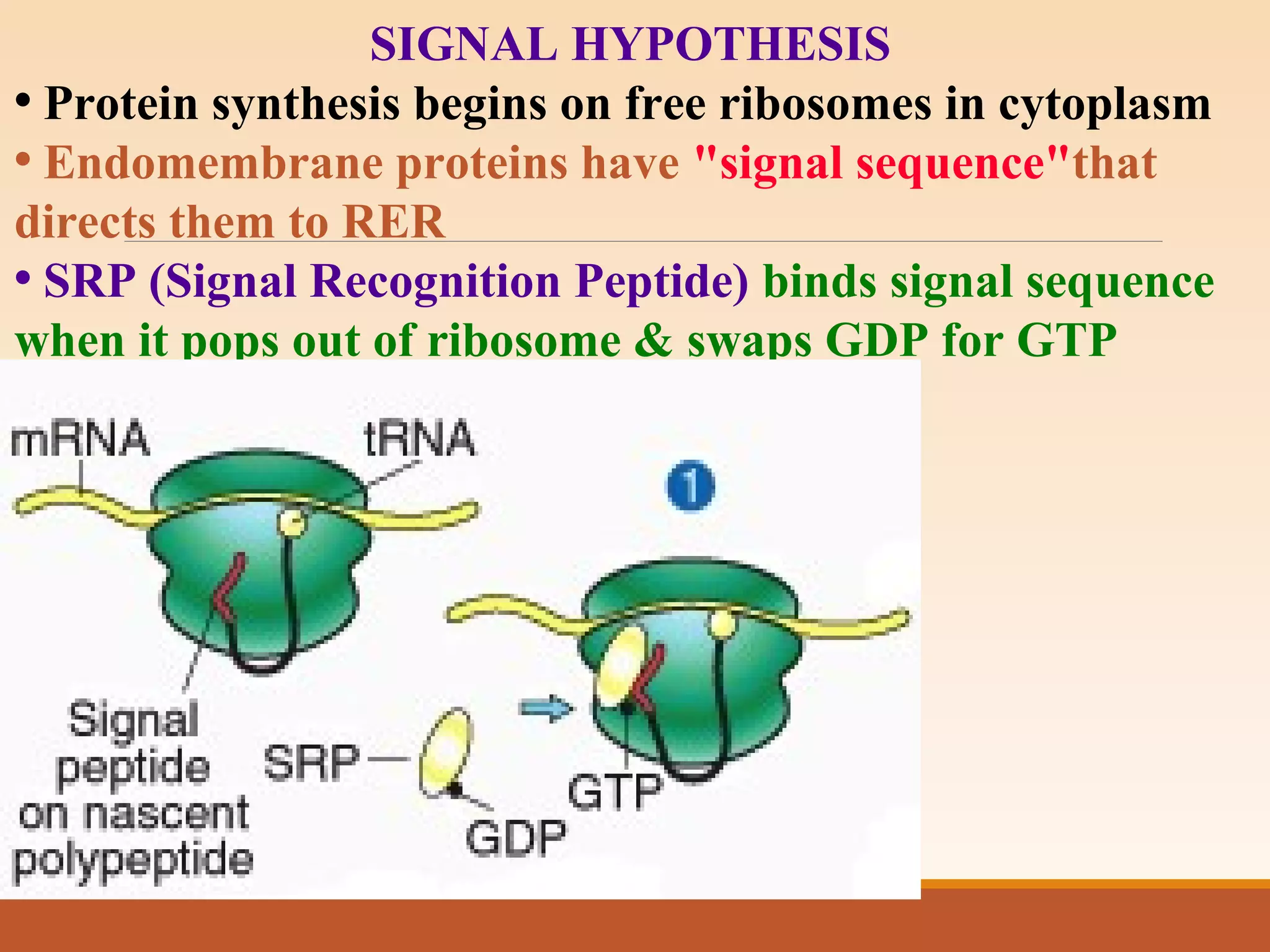
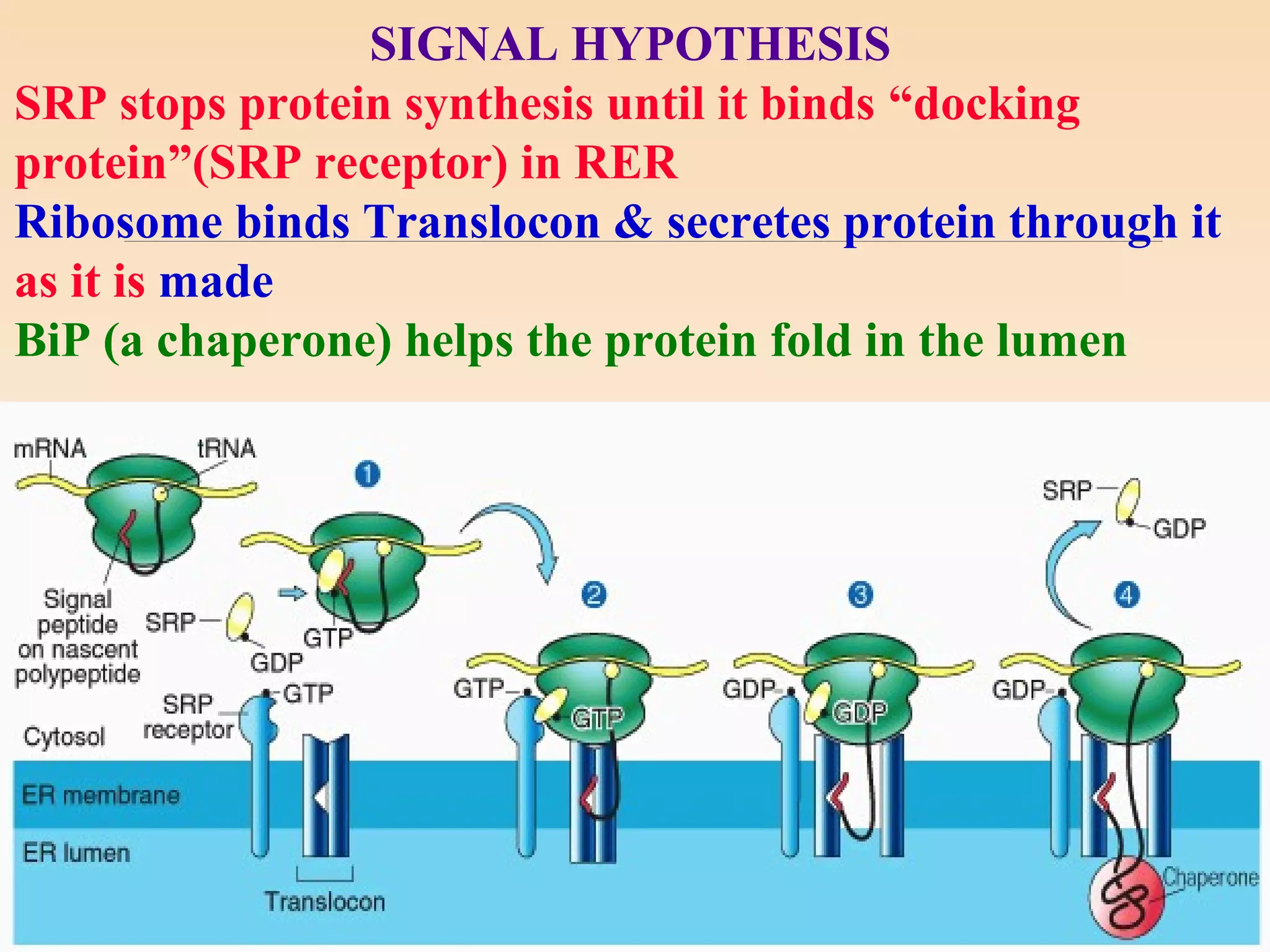
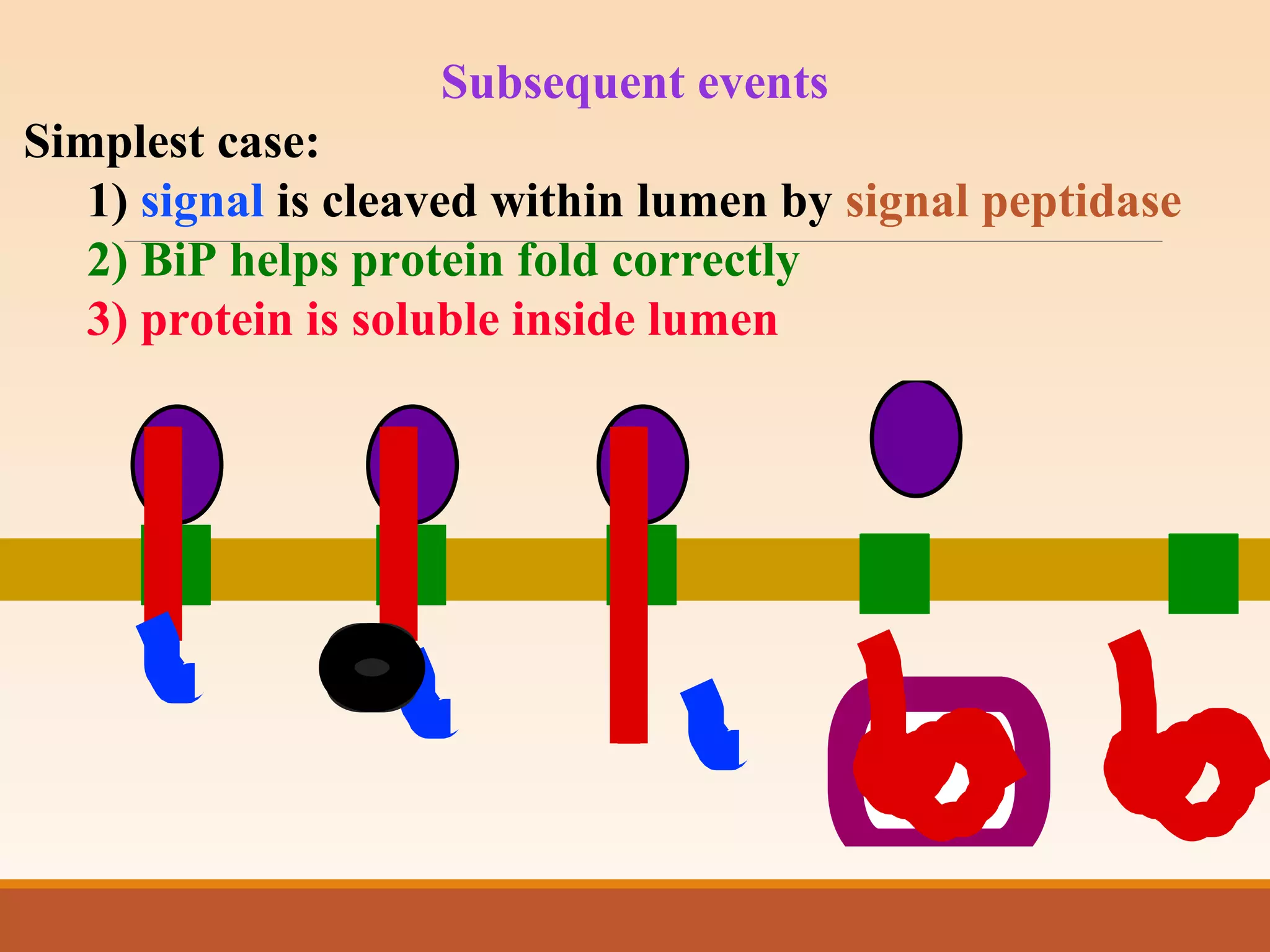
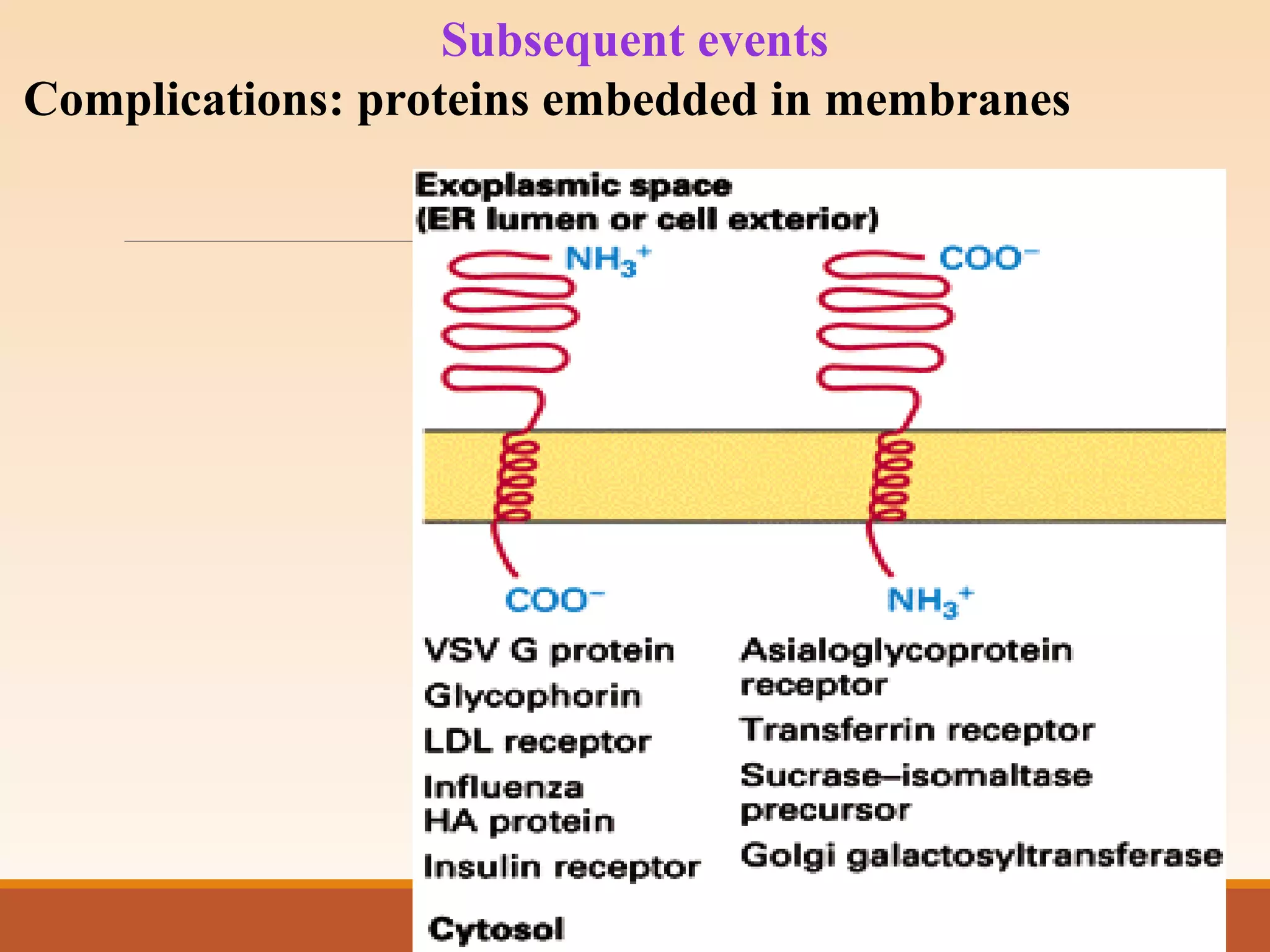
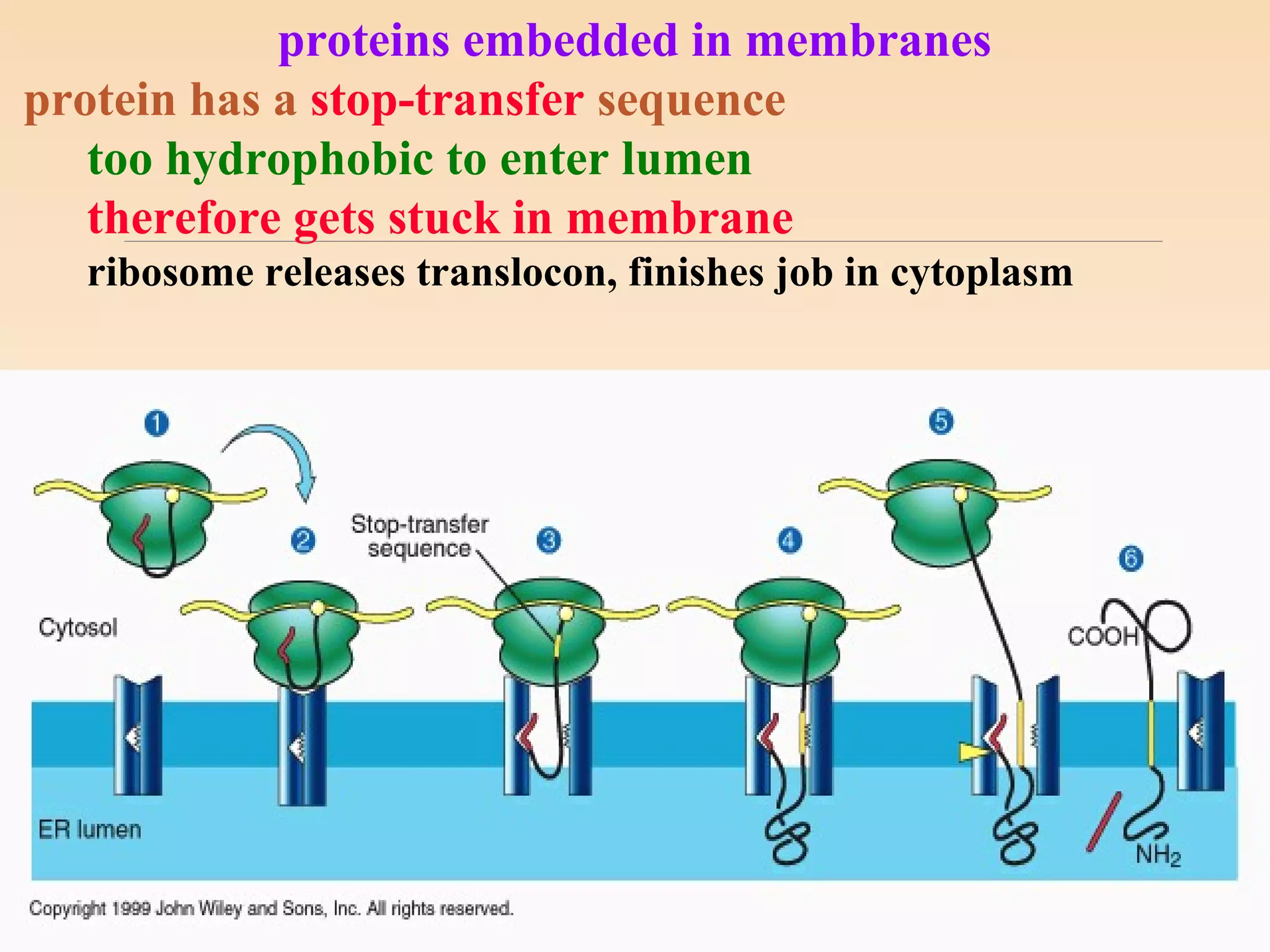
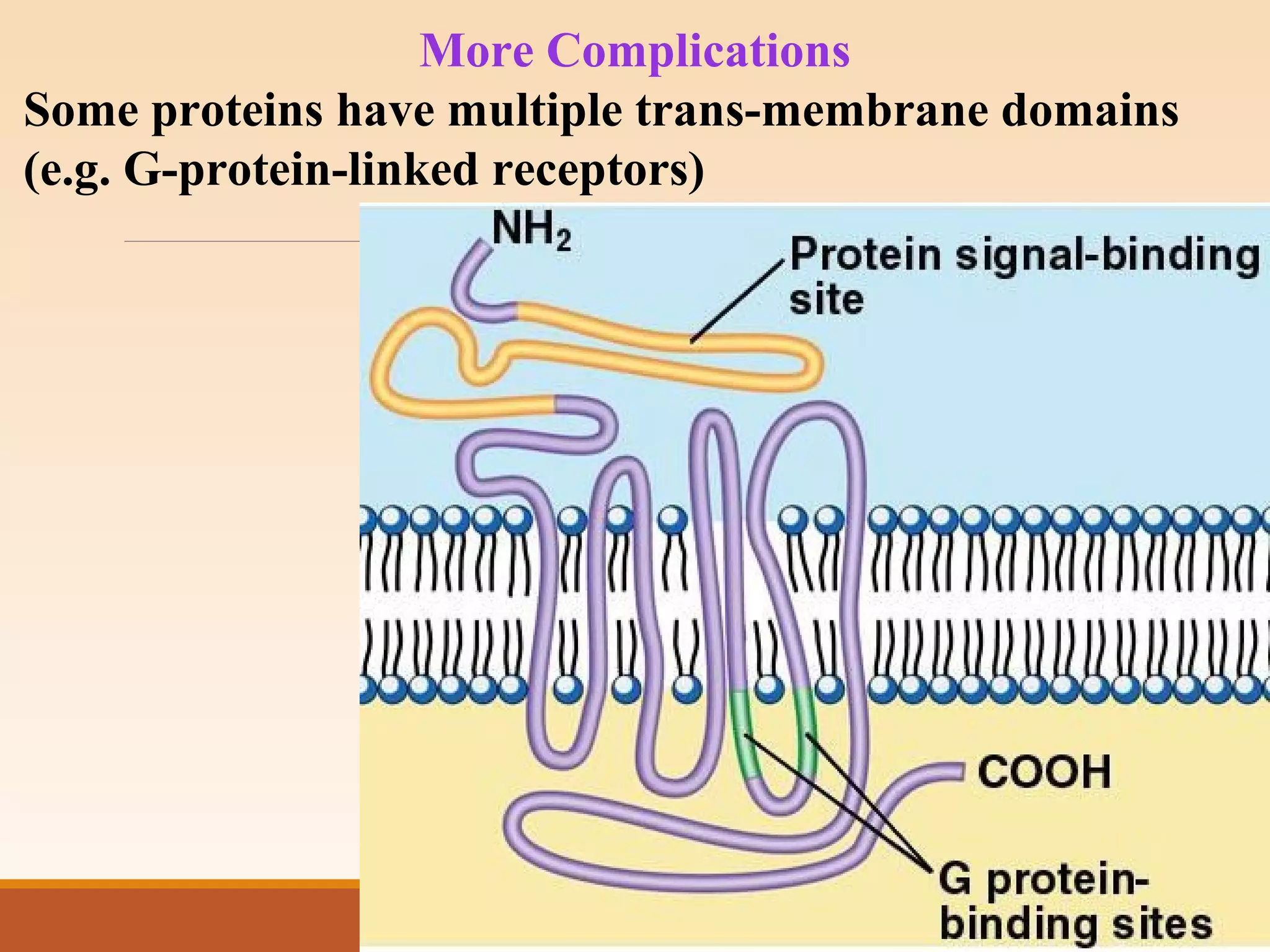
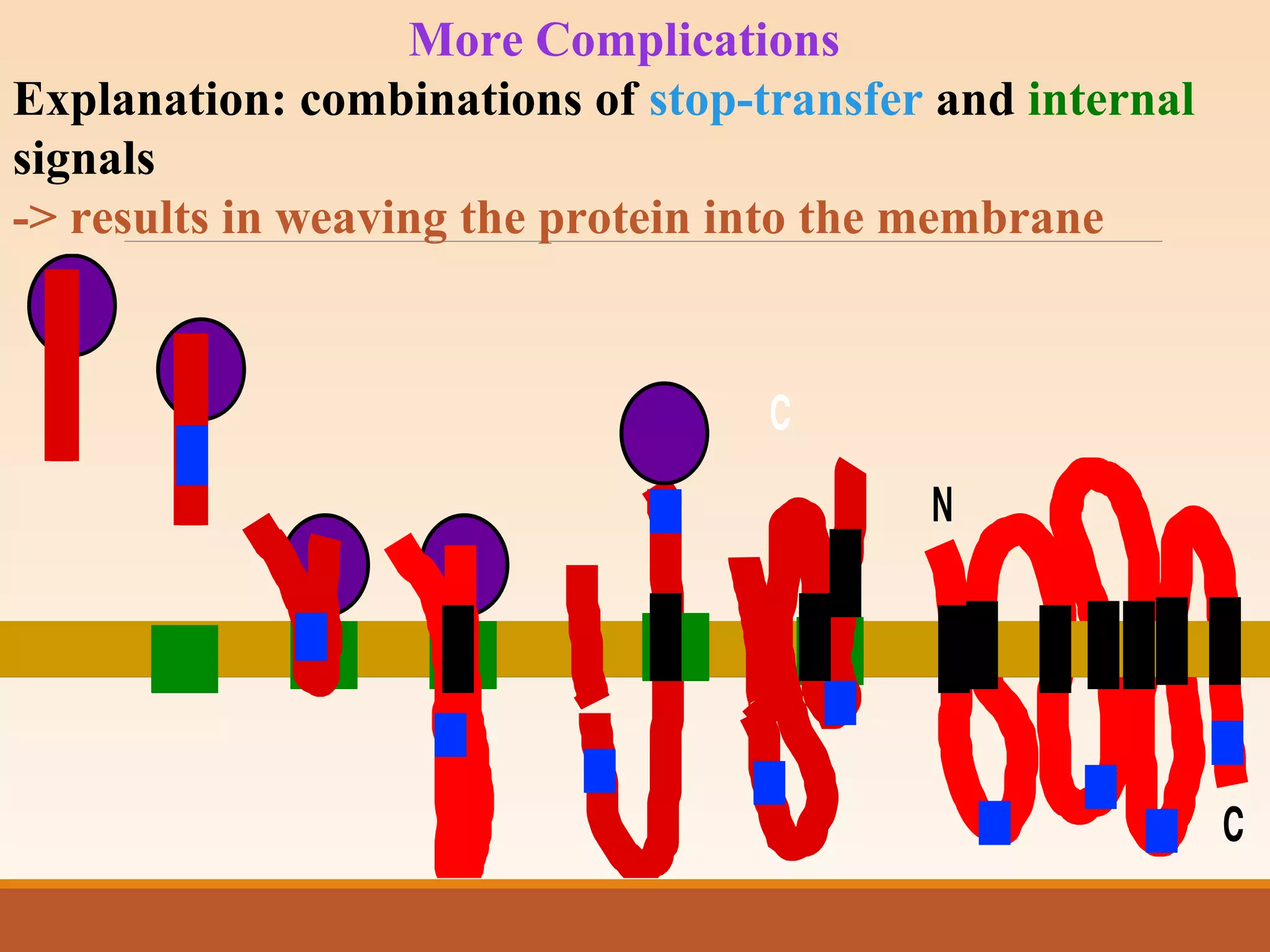
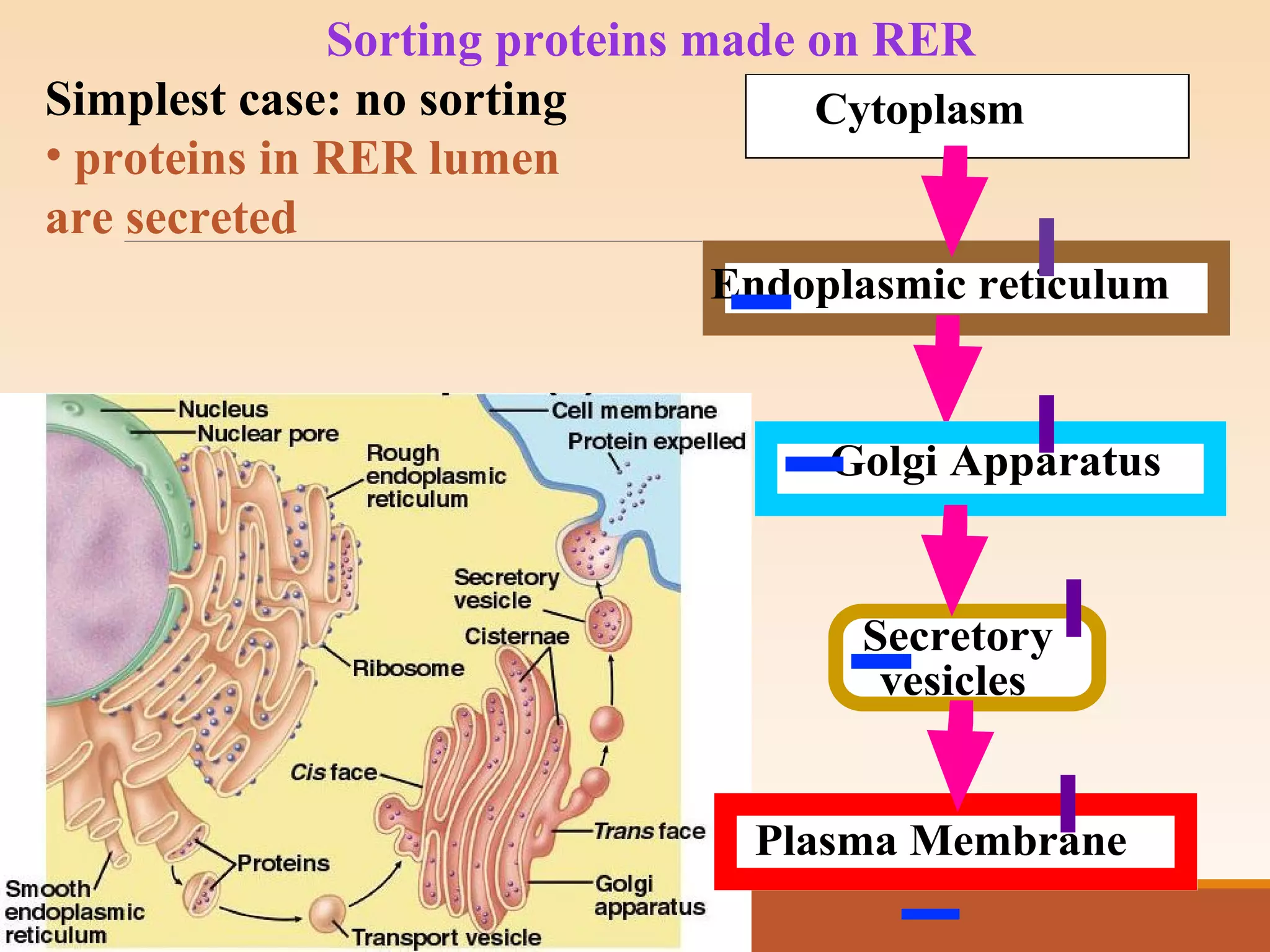
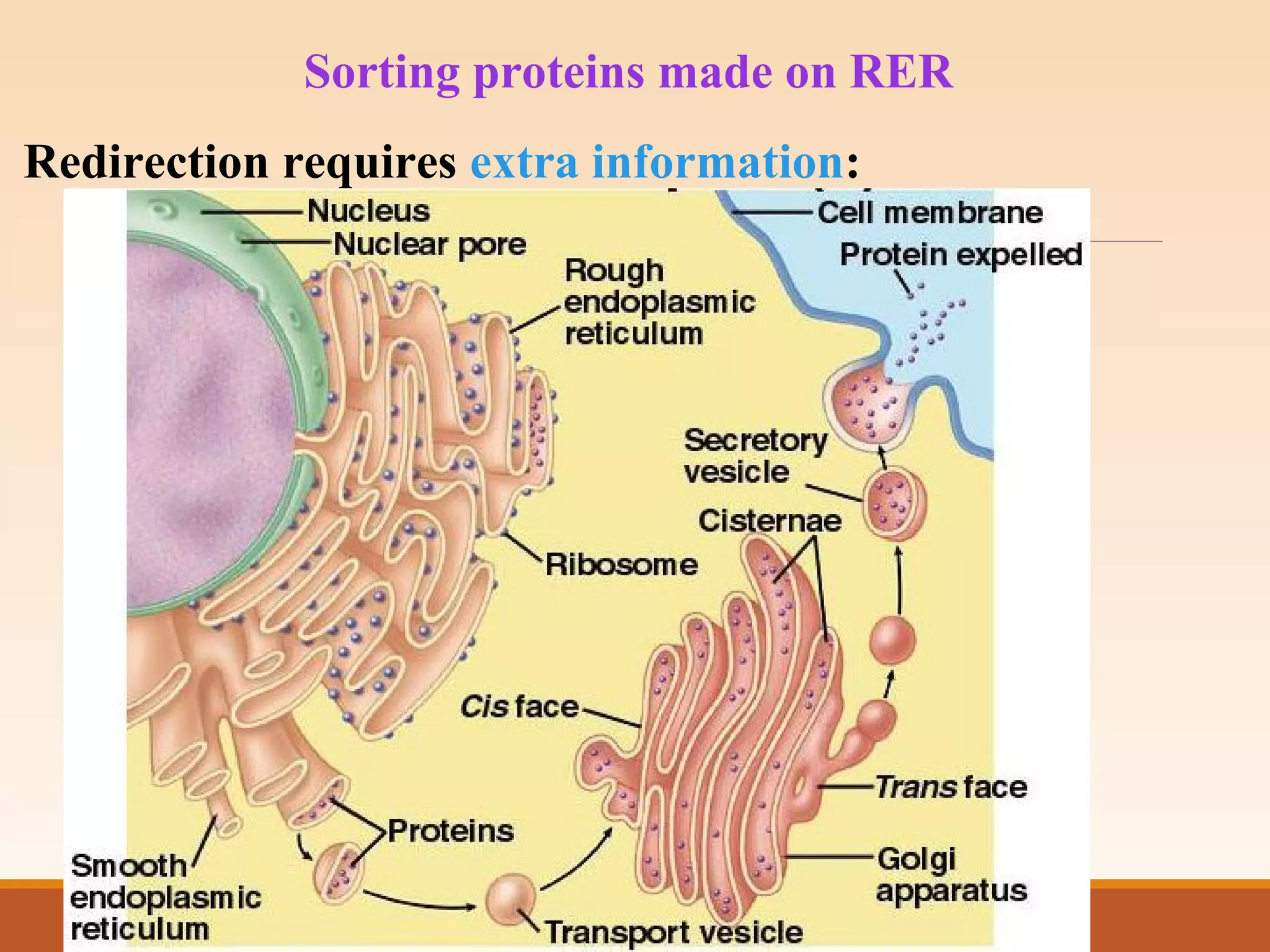
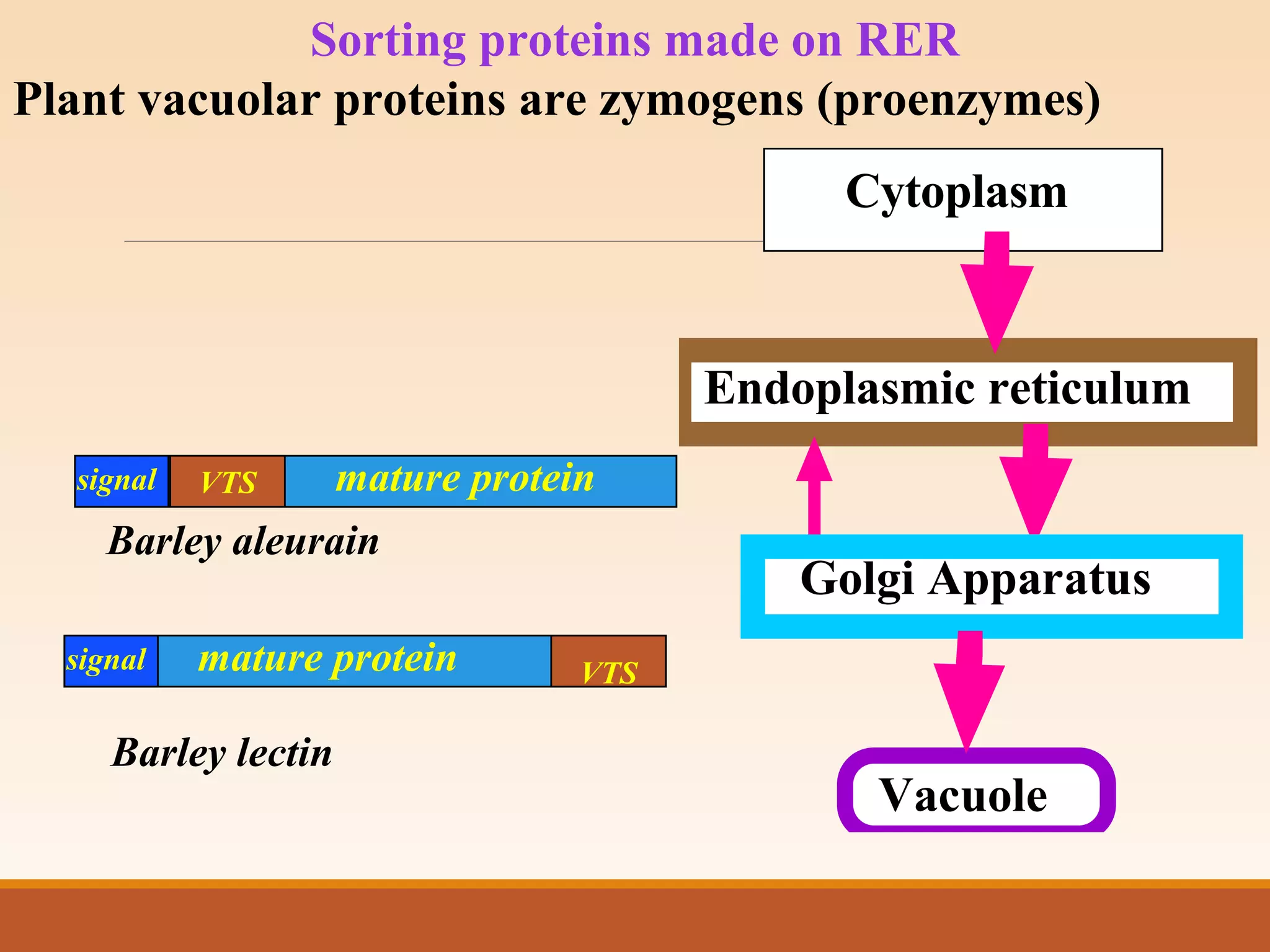
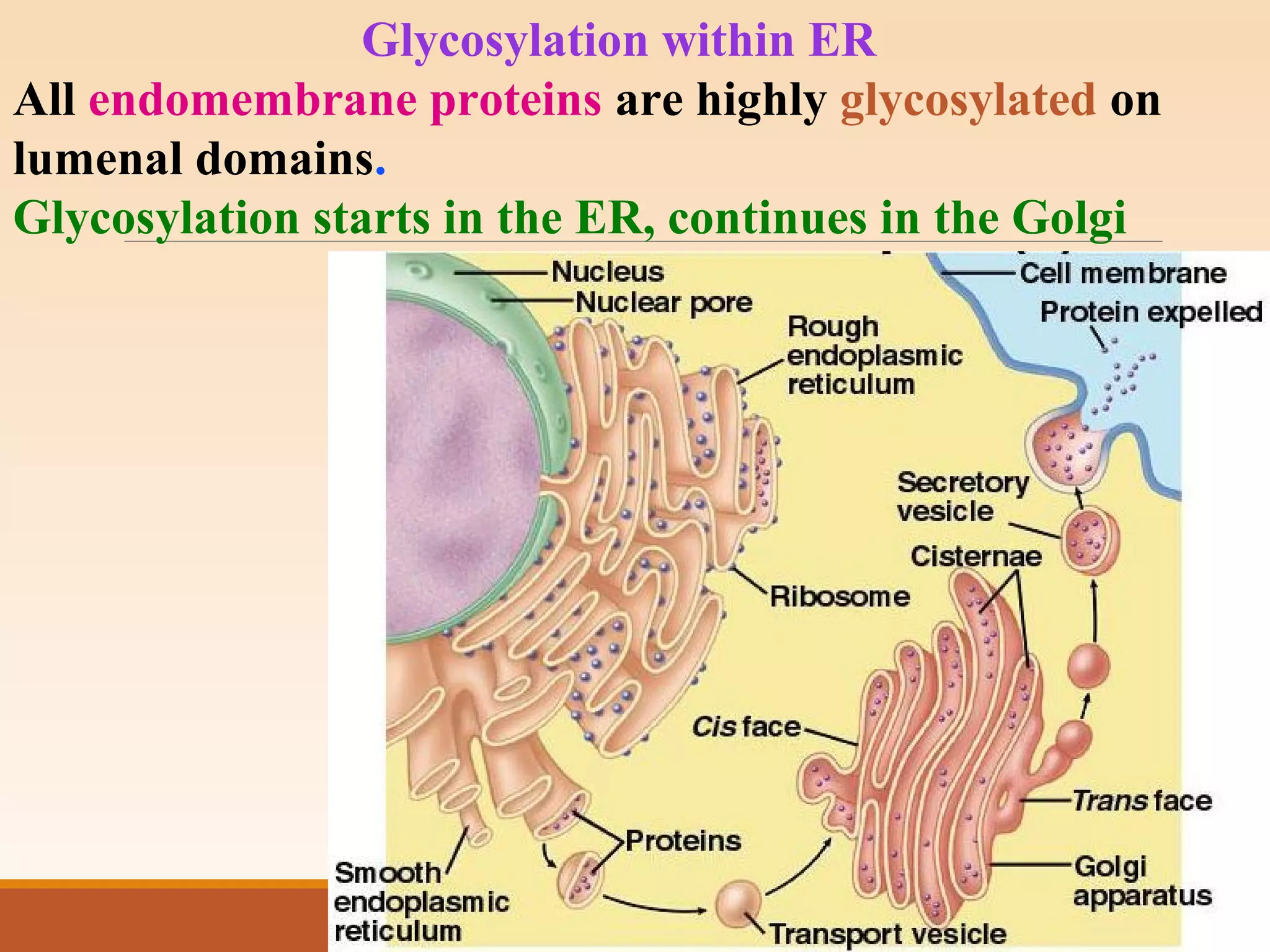
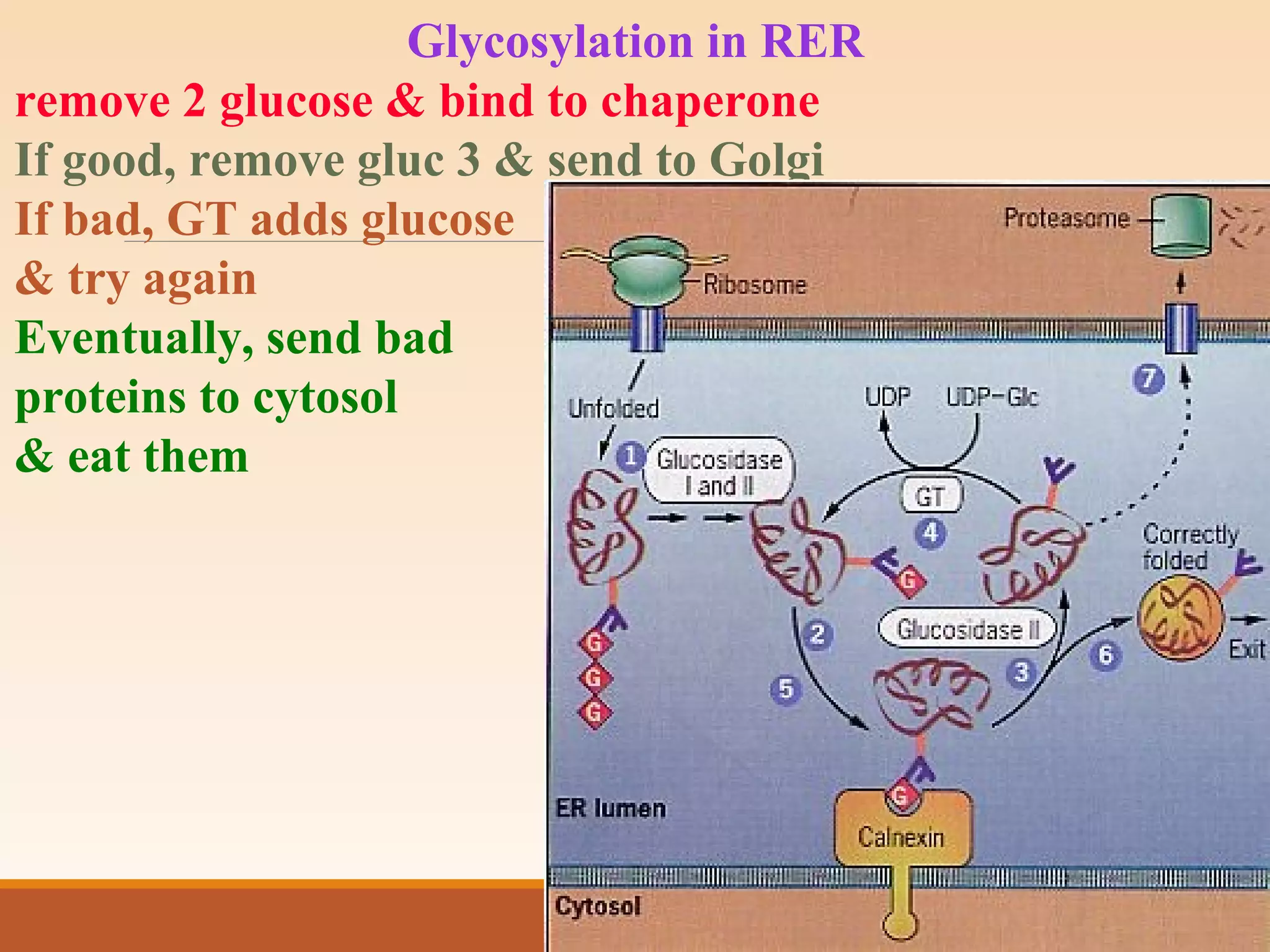
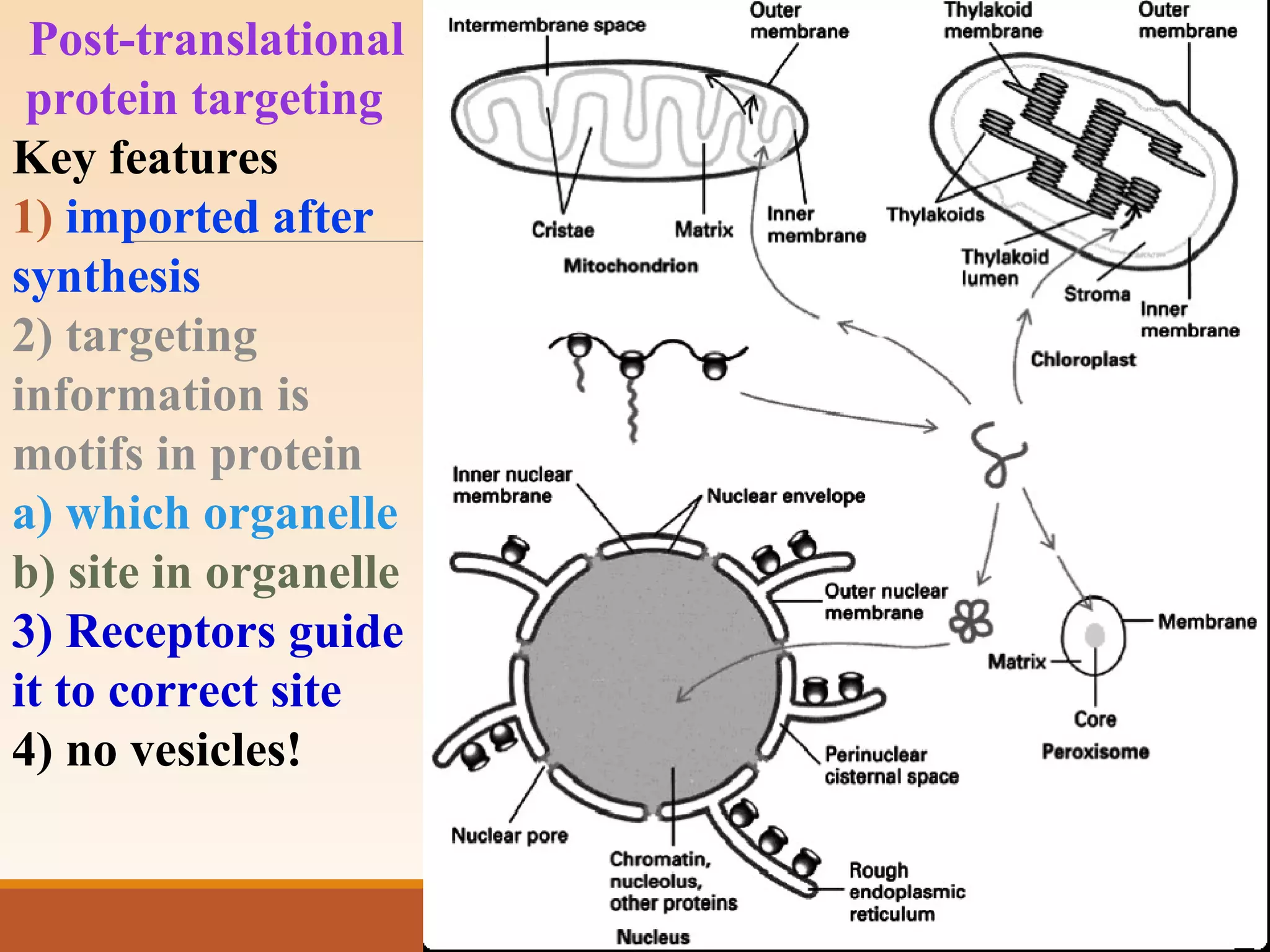
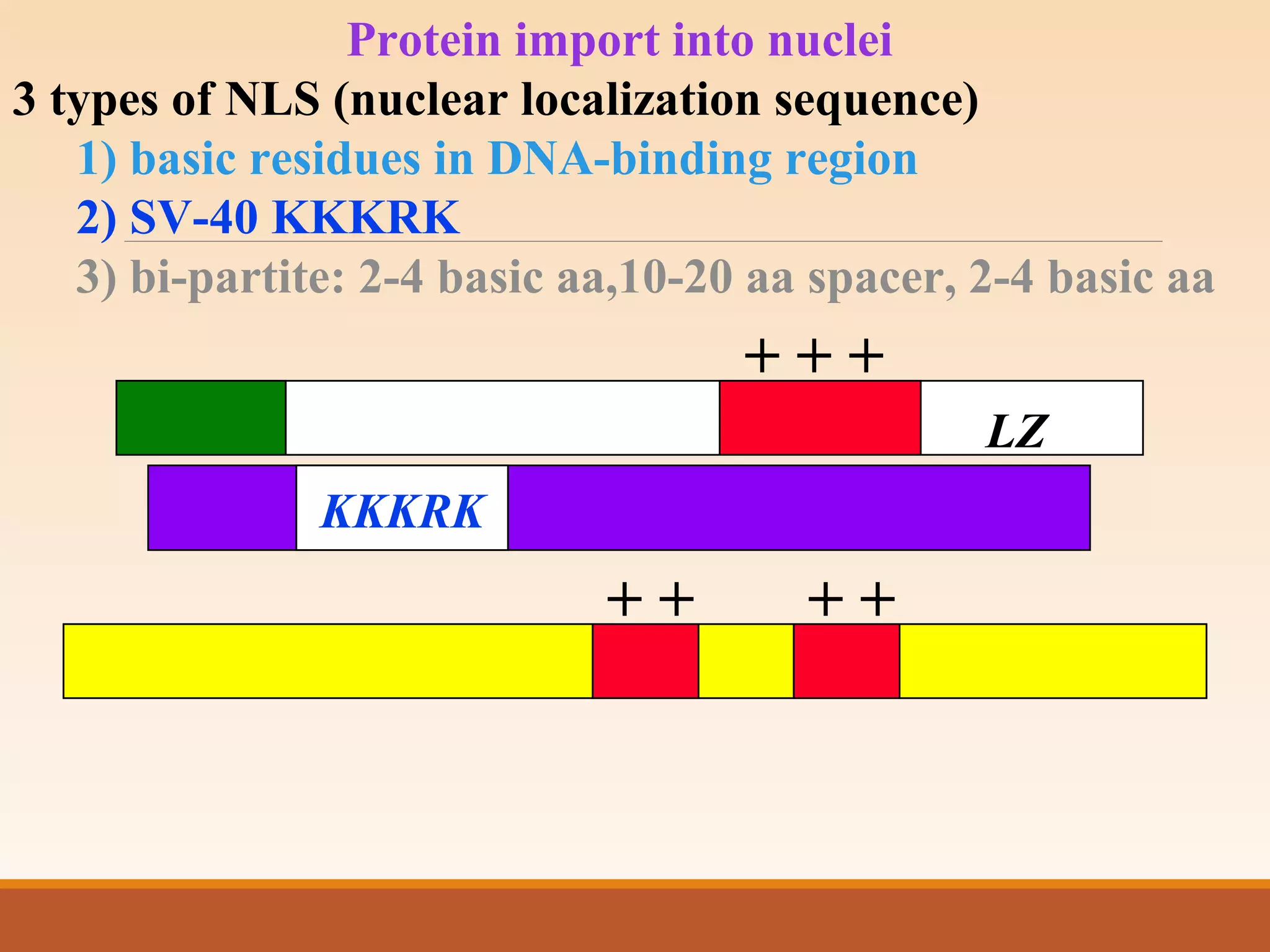
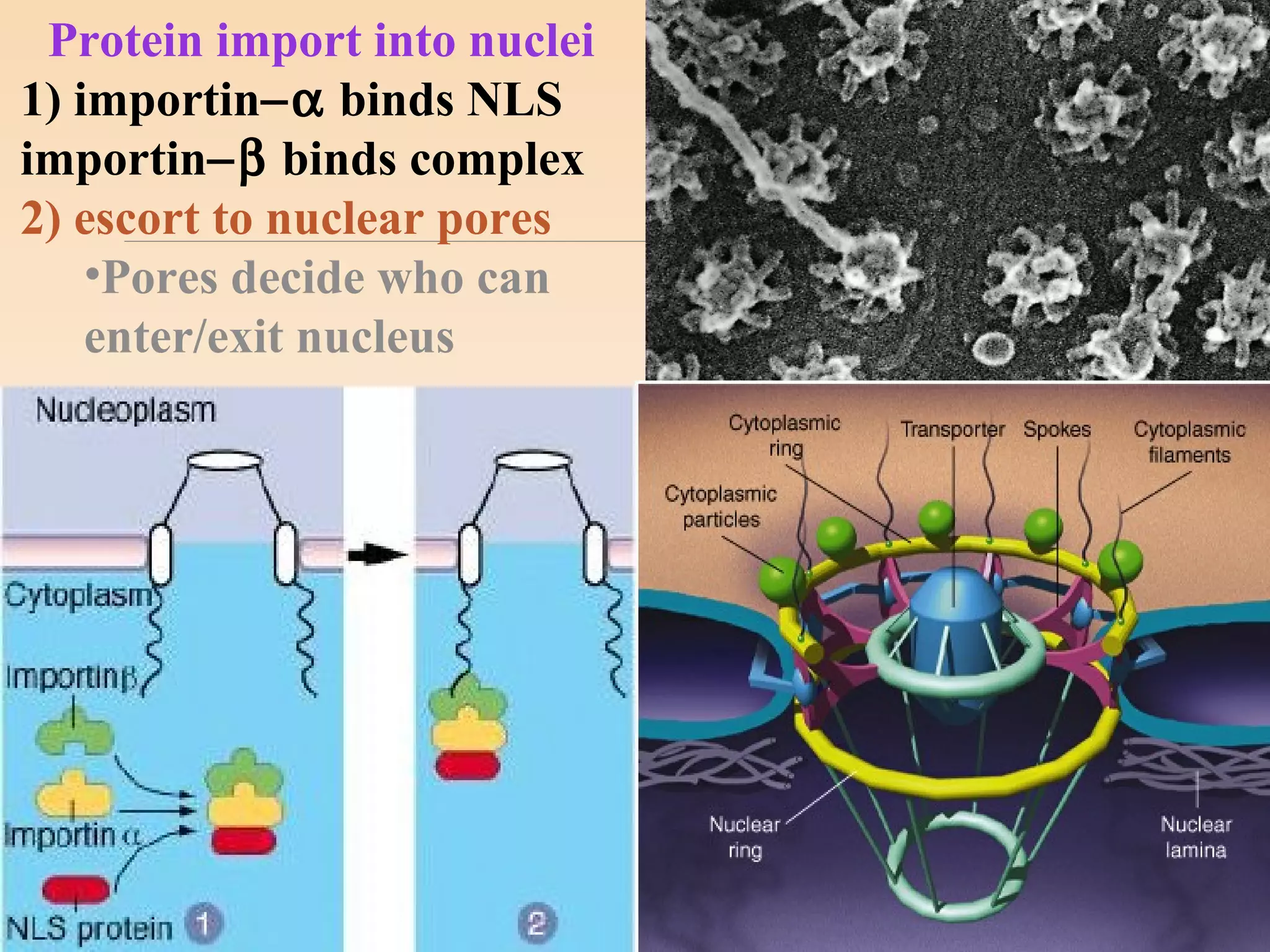
The presentation by Archana Soni discusses protein targeting pathways, emphasizing post-translational and co-translational mechanisms, where proteins are synthesized on free ribosomes and directed to their destinations via signal sequences. Key topics include the role of chaperones, glycosylation, and the specific motifs that guide proteins to organelles such as the endoplasmic reticulum, Golgi apparatus, and lysosomes. It also covers the nuclear import process, highlighting various nuclear localization sequences and the mechanisms involved in escorting proteins into the nucleus.