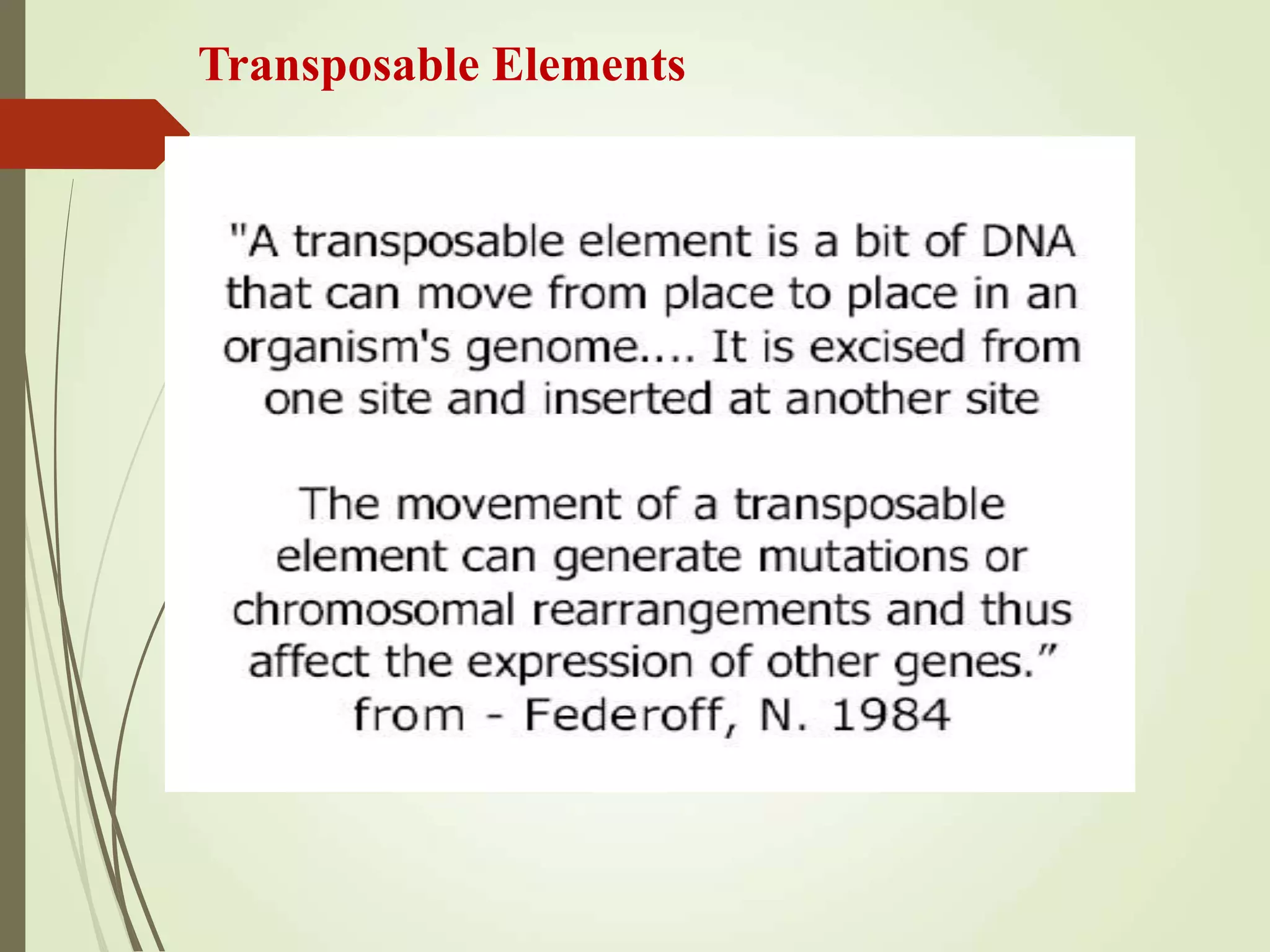
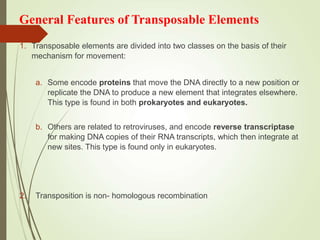
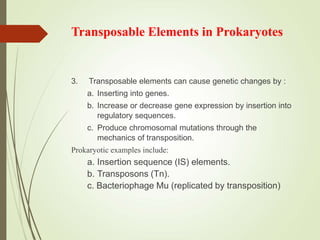
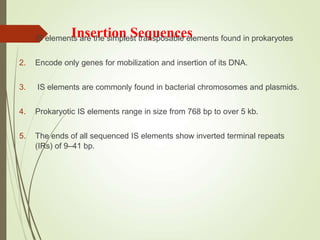
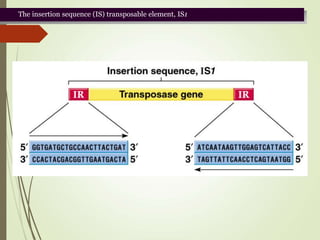
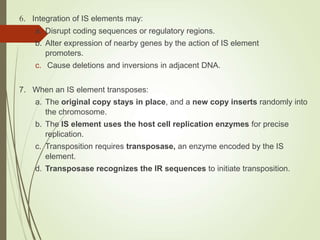
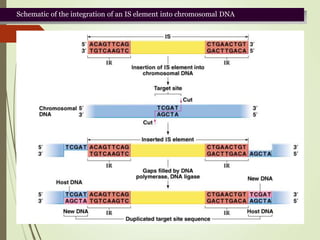
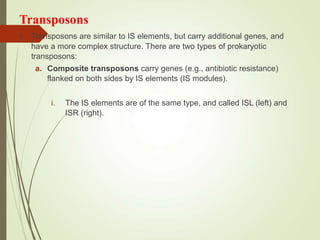
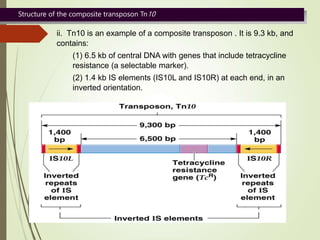
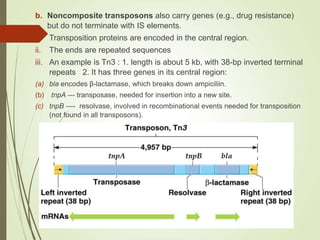
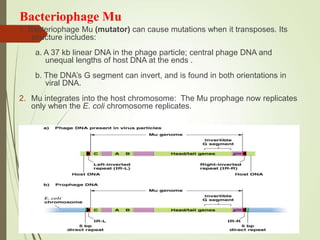
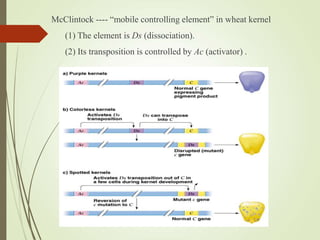
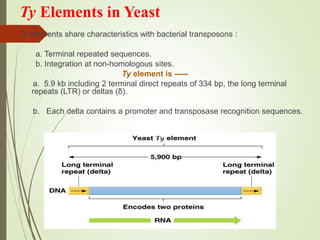
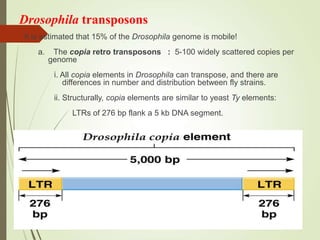
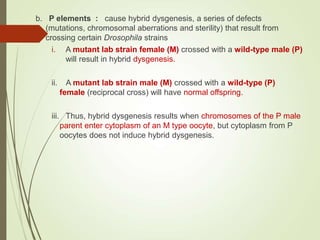
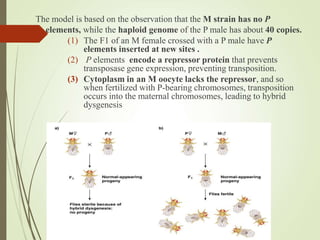
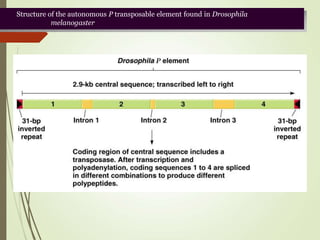
Transposable elements are segments of DNA that can change their position within the genome. They are divided into two classes based on their mechanism of movement. Class 1 elements encode reverse transcriptase and form DNA copies from RNA transcripts. Class 2 elements encode proteins that directly move the DNA. Transposable elements in prokaryotes include insertion sequences (IS elements) and transposons, while eukaryotes also contain retrotransposons similar to retroviruses. Transposition can disrupt genes or regulatory sequences and cause mutations. McClintock's work with maize introduced the concept of mobile genetic elements controlling gene expression.