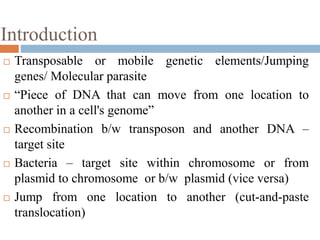
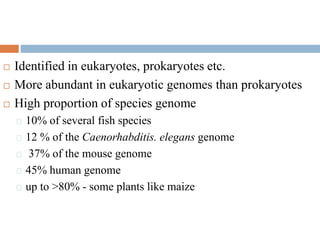
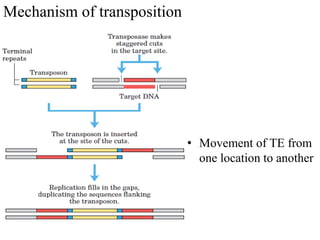
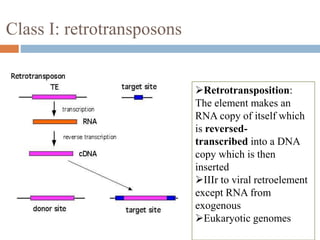
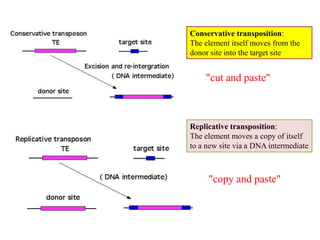
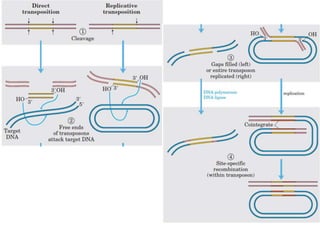
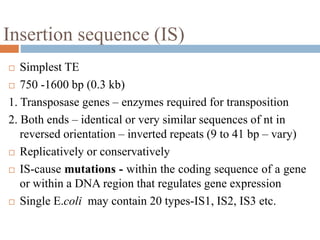
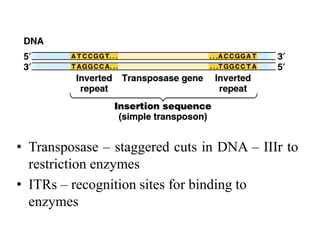
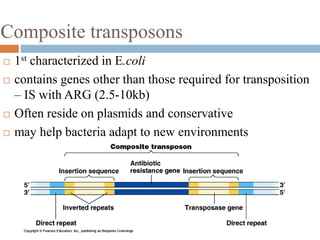
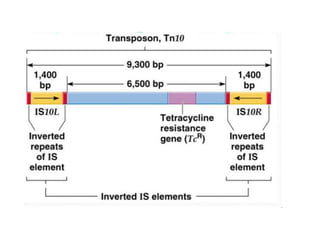
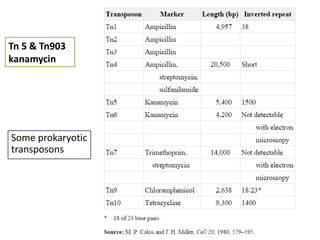
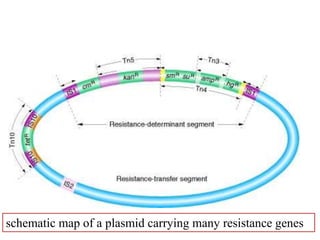
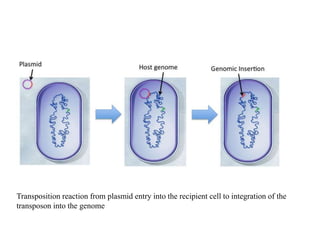
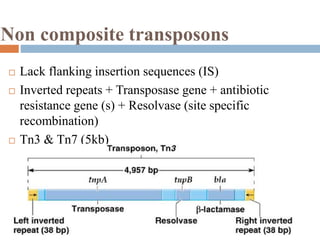
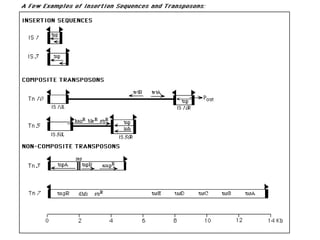
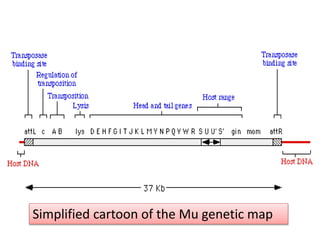
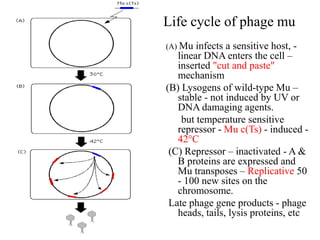
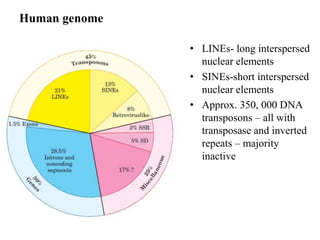
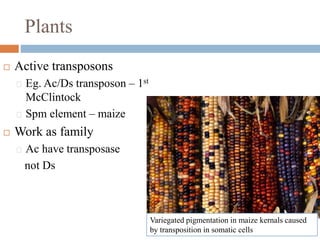
This document provides an overview of transposable elements (TEs). It discusses that TEs are DNA sequences that can move within genomes. There are two major classes of TEs - retrotransposons which move via an RNA intermediate, and DNA transposons which move directly through a cut-and-paste or copy-and-paste mechanism. The document outlines different types of TEs found in bacteria, eukaryotes, and the human genome. It also discusses the impact of TEs in causing mutations and their applications in genetic engineering and mutagenesis experiments.