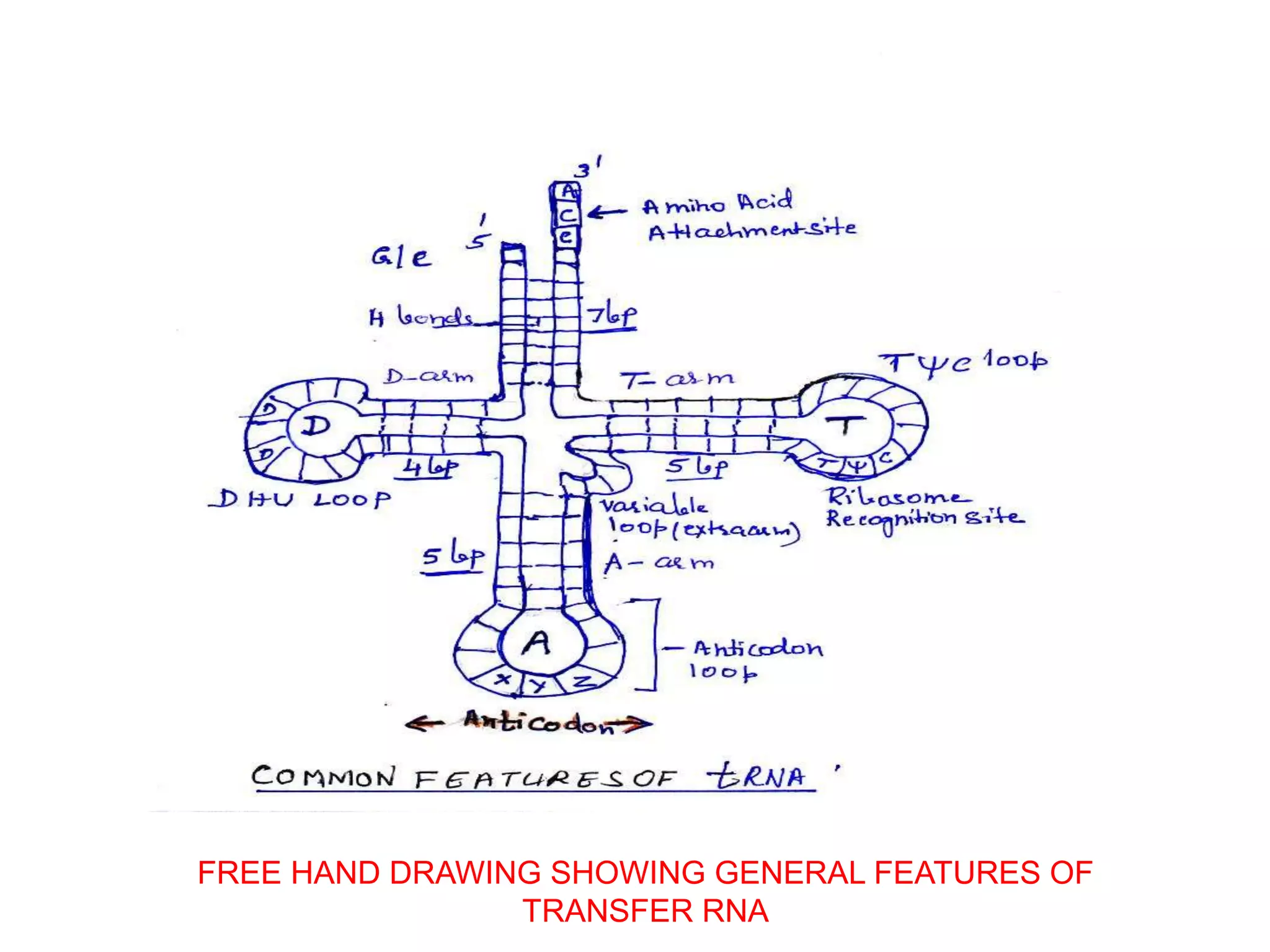
The document discusses transfer RNA (tRNA), a crucial molecule in protein synthesis that carries amino acids to ribosomes. It highlights the structure and function of tRNA, including its cloverleaf configuration, anticodon pairings, and the process of aminoacylation, where specific amino acids are attached to tRNA by aminoacyl-tRNA synthetases. Additionally, it covers the transcription and post-transcriptional modifications of tRNA in both eukaryotic and prokaryotic cells.