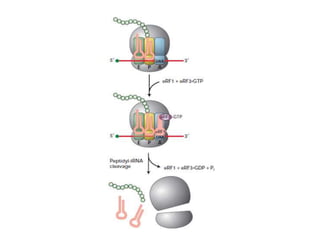
This document discusses the process of translation in three sentences:
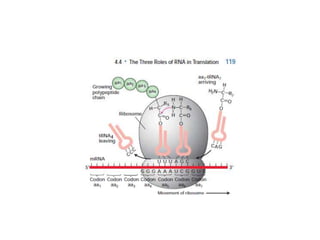
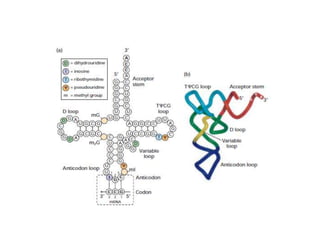
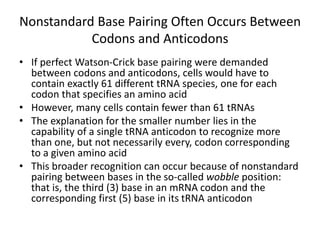
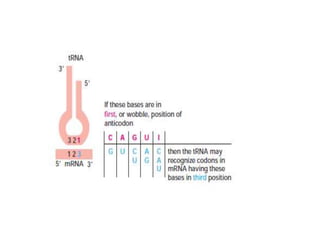
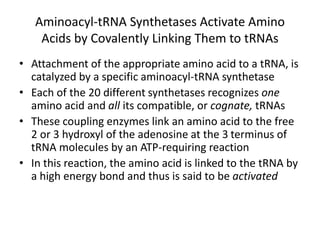
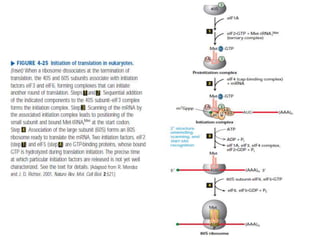
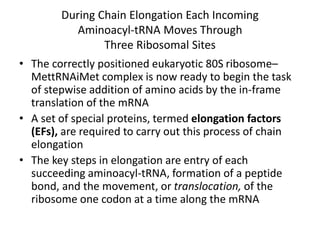
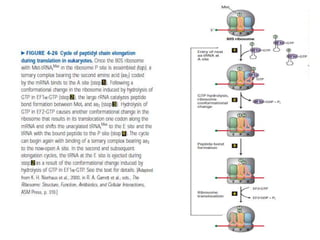
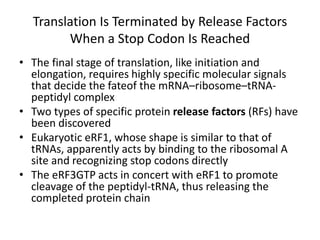
Messenger RNA carries genetic information from DNA in the form of codons that specify amino acids. Transfer RNA matches codons to their corresponding amino acids through complementary base pairing between its anticodon and the mRNA codon. Ribosomes catalyze the formation of peptide bonds between amino acids specified by mRNA and delivered by tRNA to assemble polypeptide chains according to the genetic code.