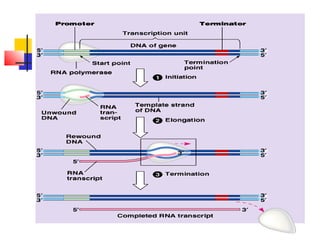
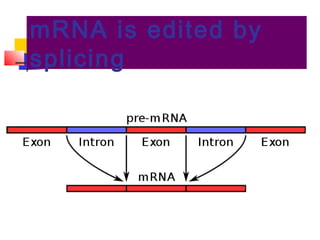
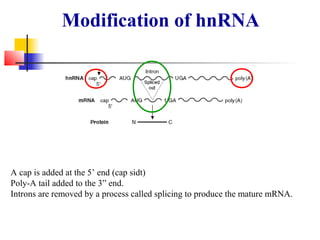
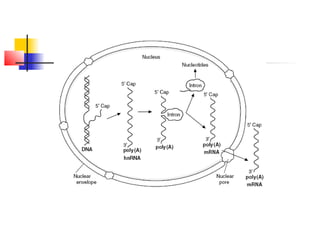
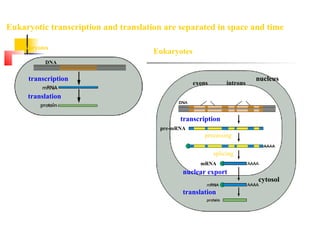
The document discusses the processes of transcription and post-transcriptional modification in RNA synthesis. It describes the roles of different types of RNA (mRNA, rRNA, tRNA) and details the stages of transcription such as initiation, elongation, and termination, as well as the modifications that occur in eukaryotic cells. Key differences between prokaryotic and eukaryotic transcription are highlighted, including the necessity for processing hnRNA into mature mRNA in eukaryotes.