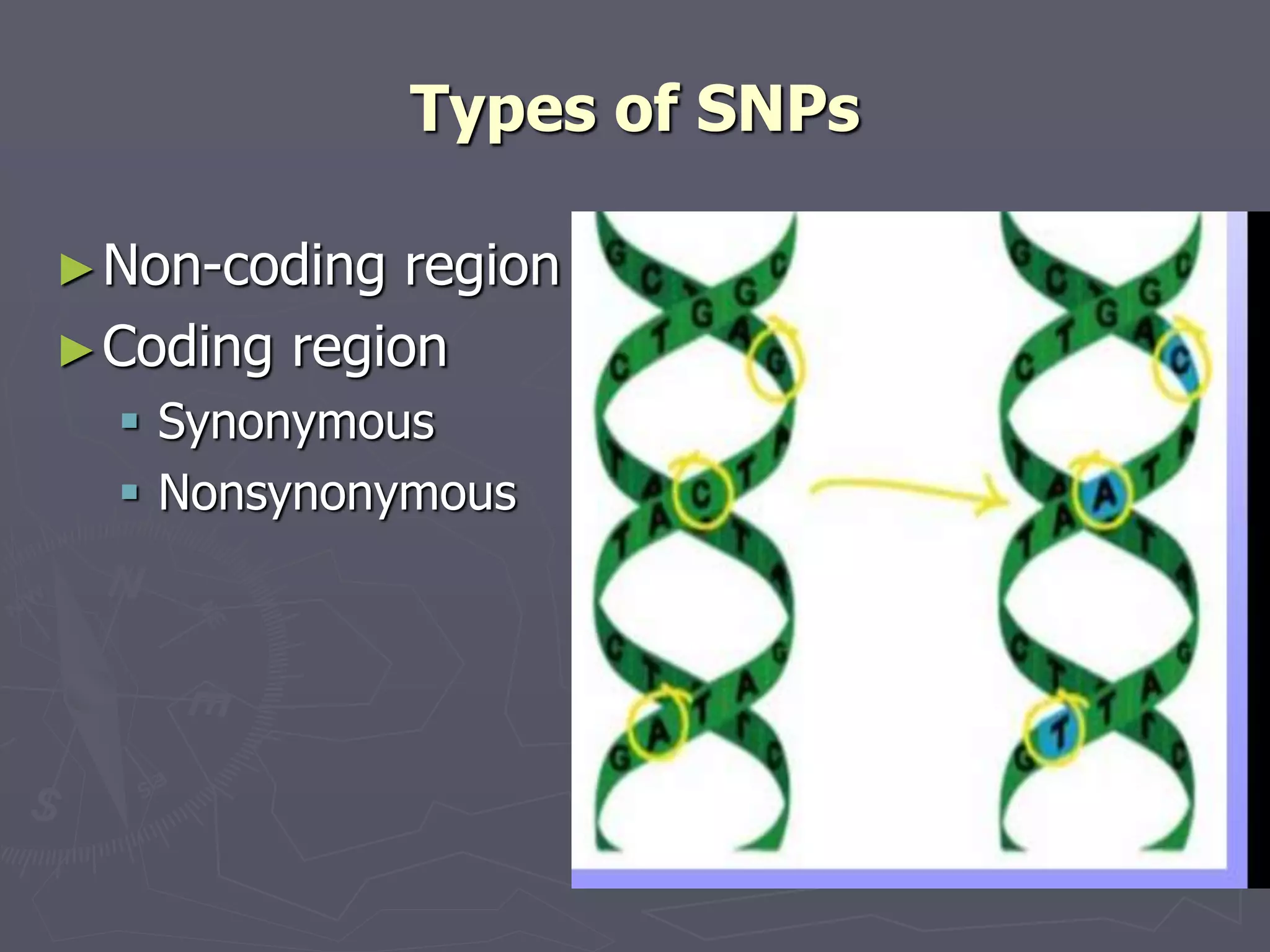
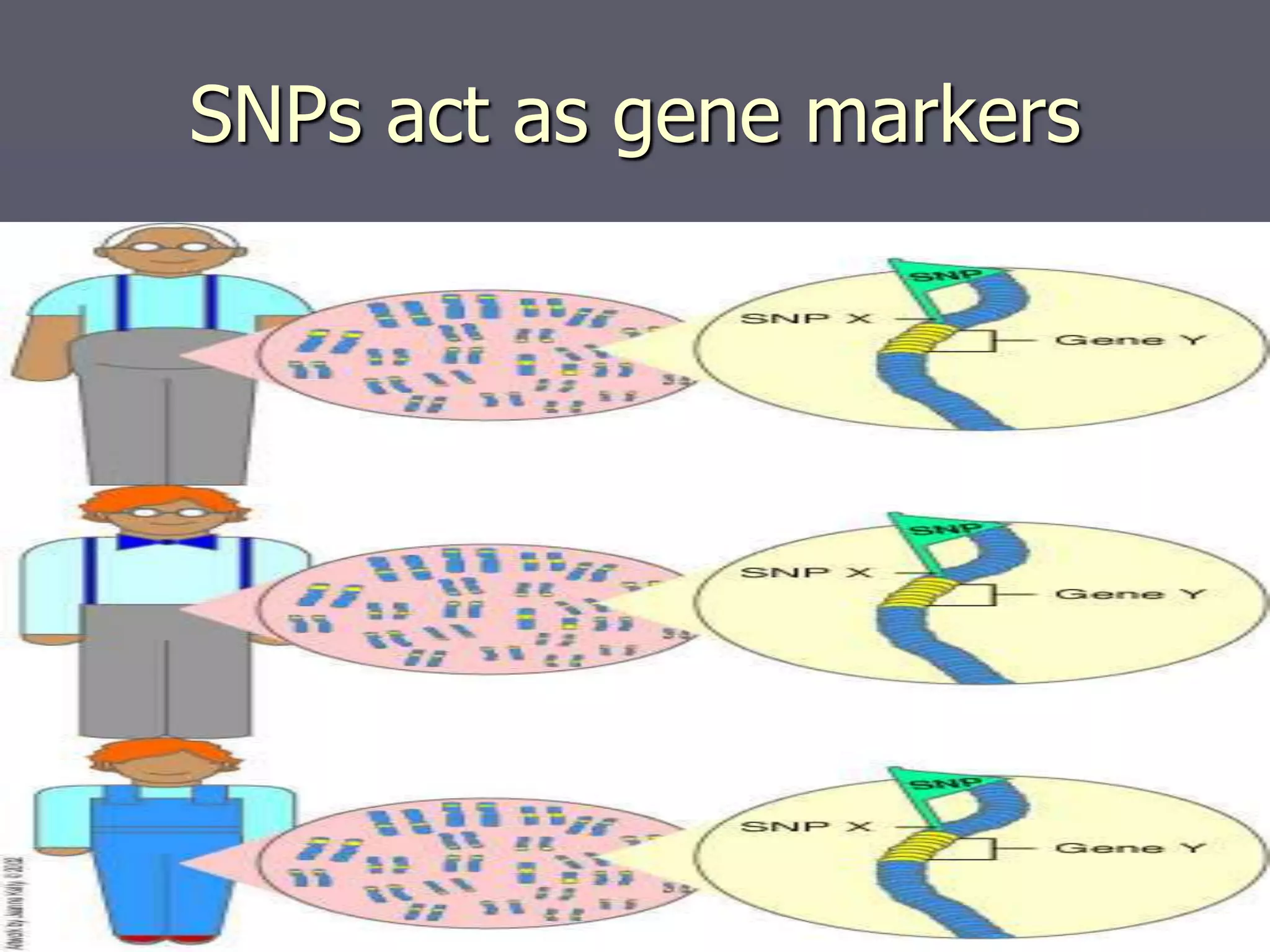
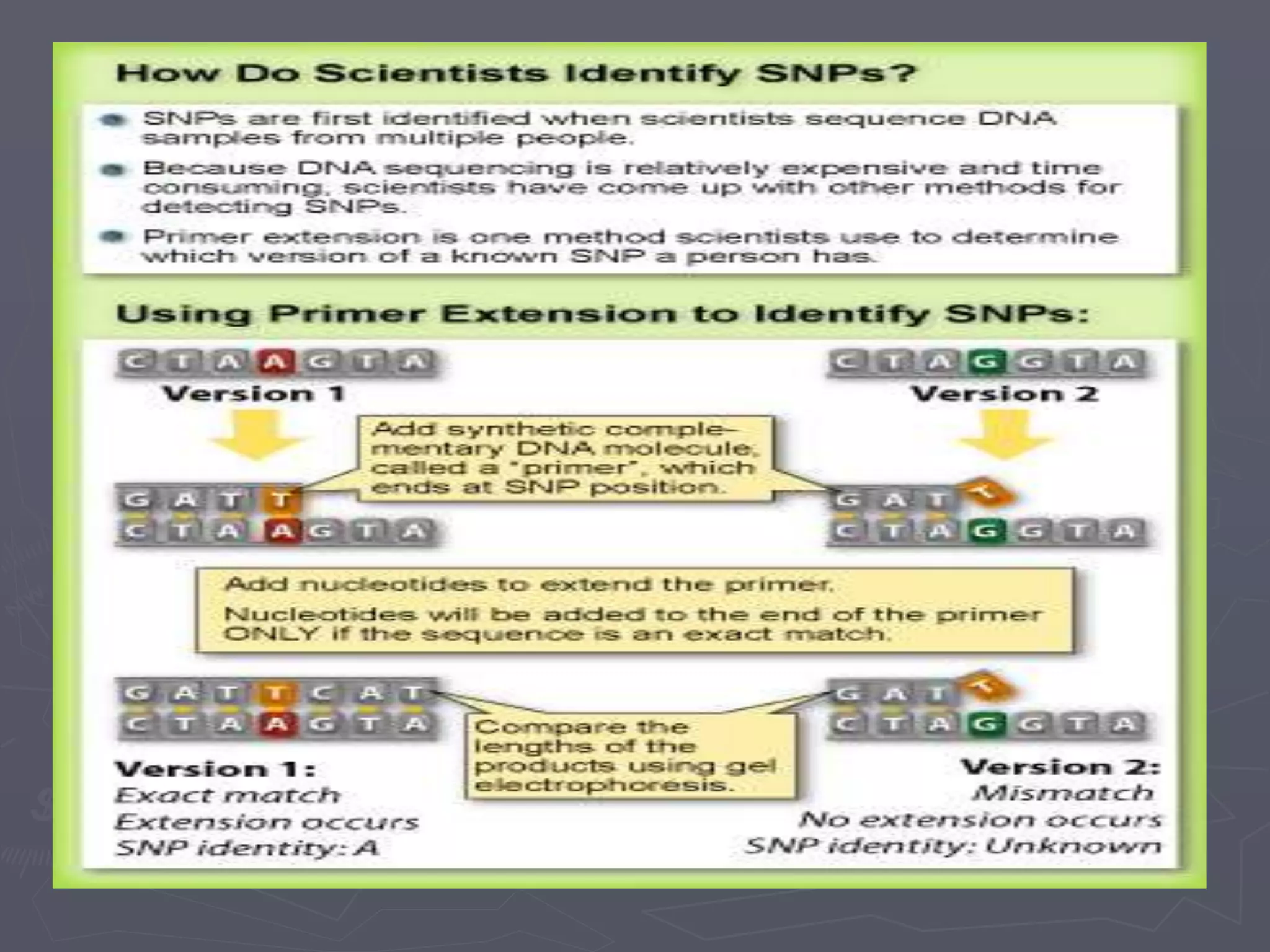
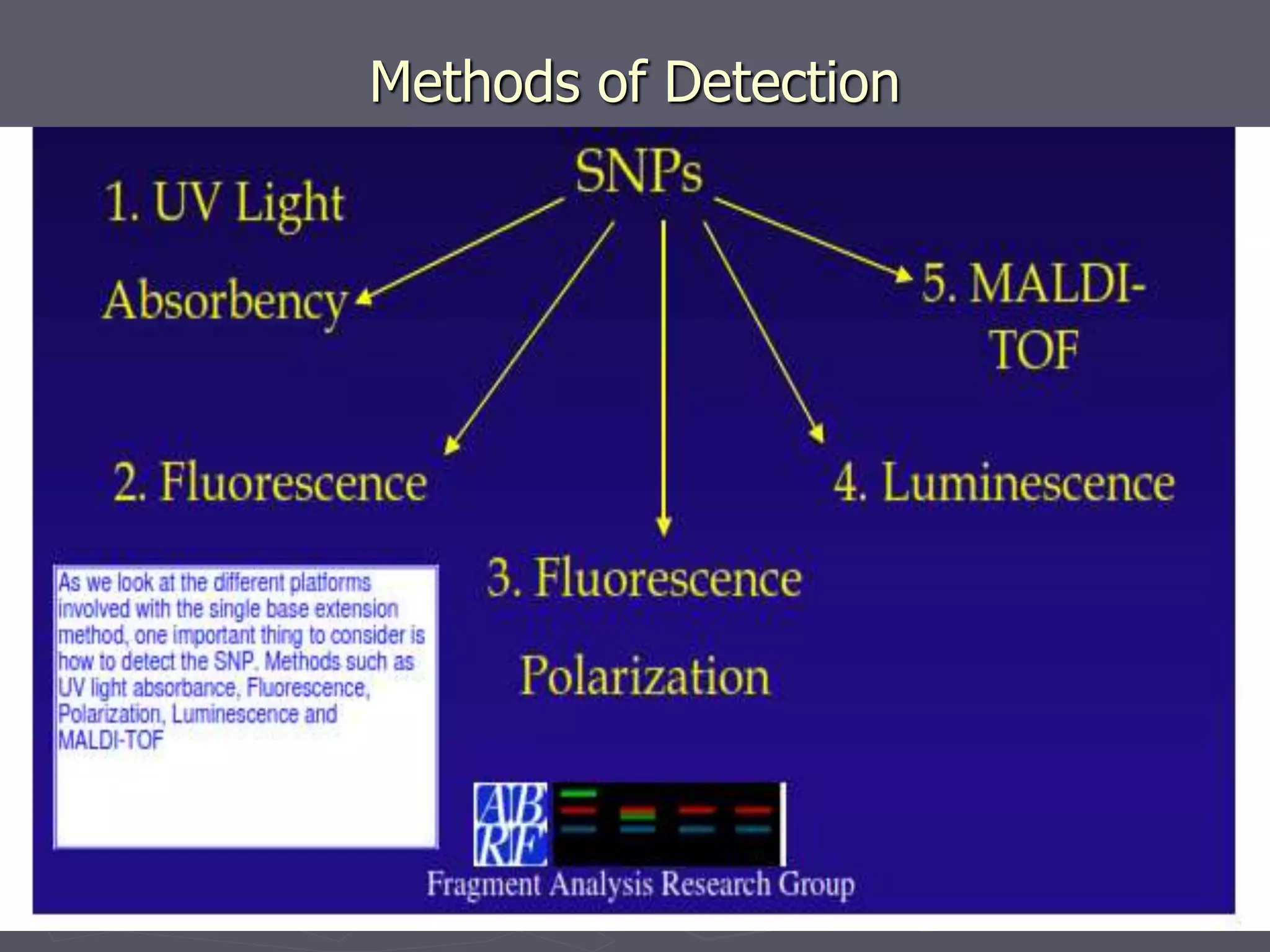
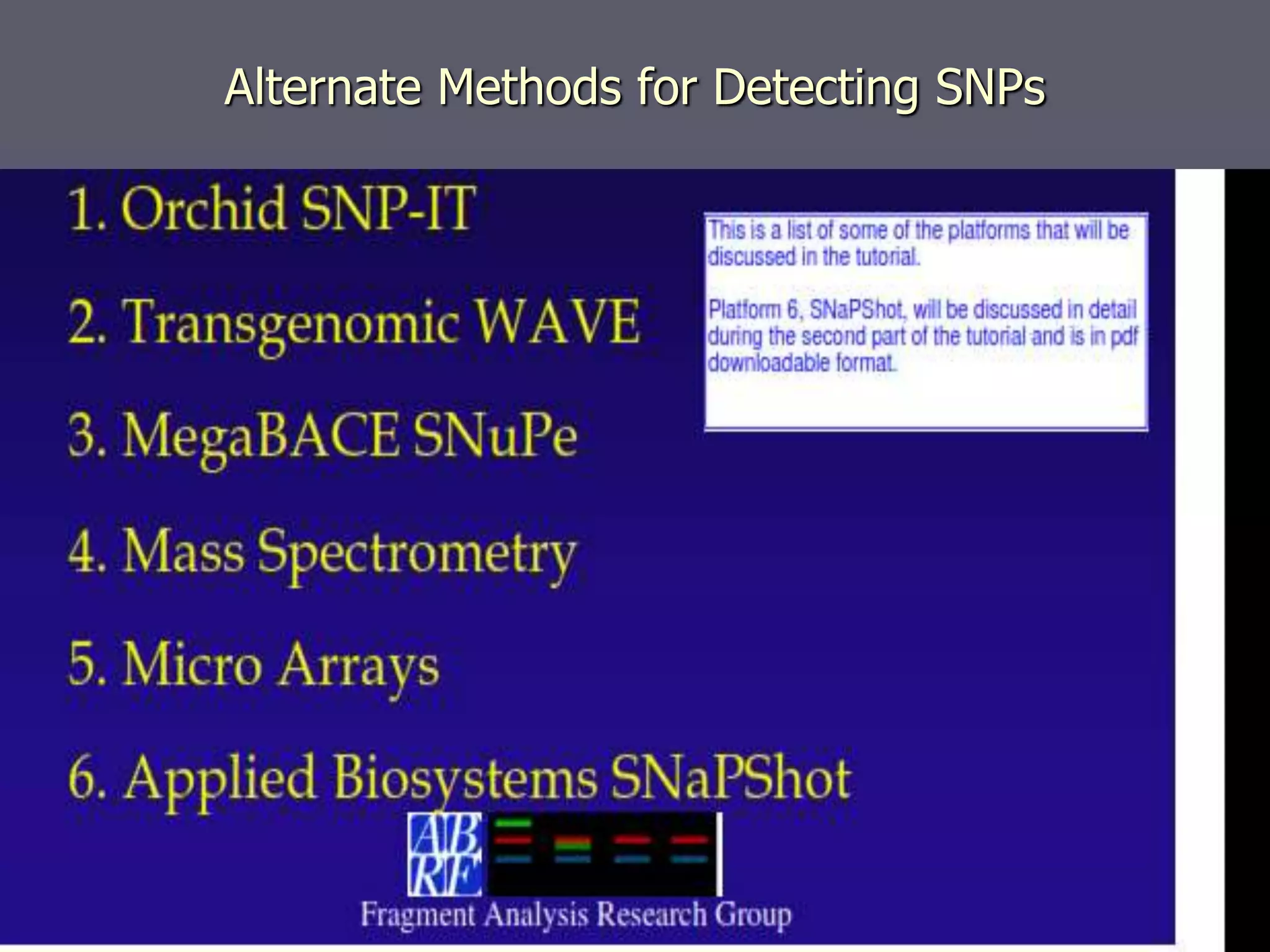
The document provides a comprehensive overview of single nucleotide polymorphisms (SNPs), including their definition, significance, variations, and techniques for detection. It discusses the methods of SNP genotyping, such as allelic specific cleavage and microchip analysis, and highlights the importance of SNPs in disease diagnosis and drug development. Additionally, various public SNP database resources are mentioned, along with recent advancements in detection methods.