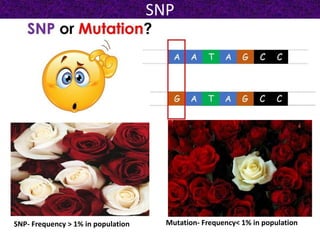
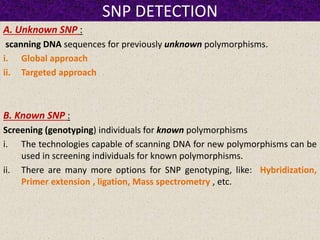
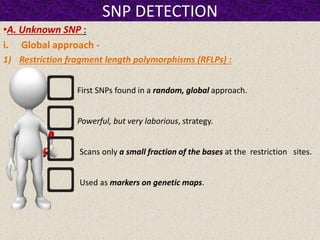
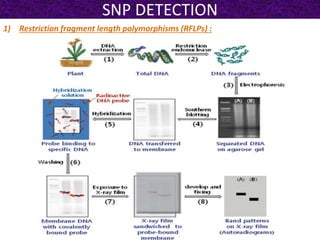
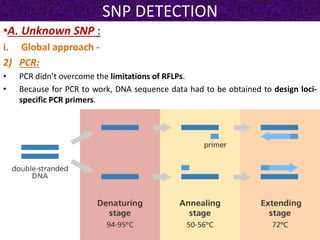
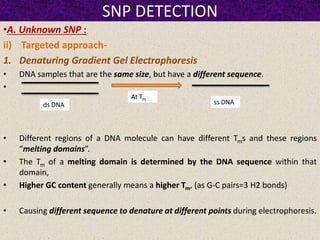
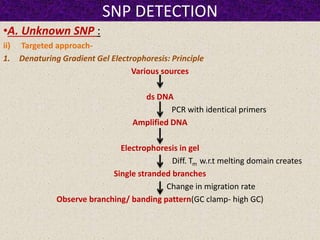
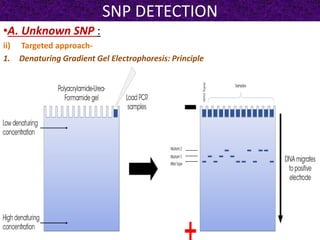
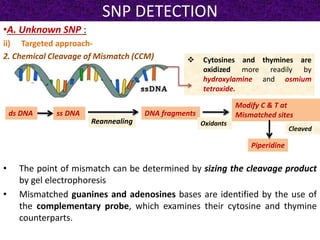
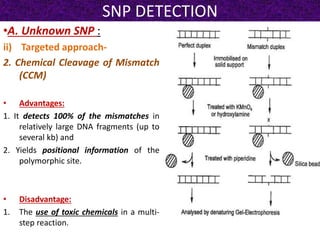
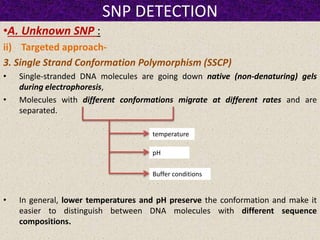
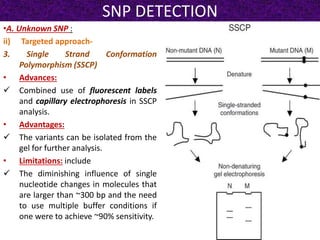
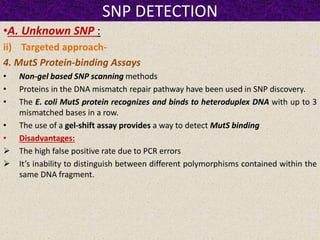
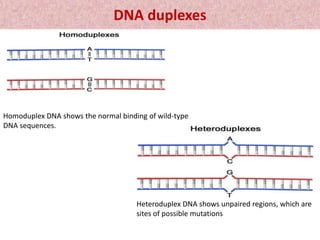
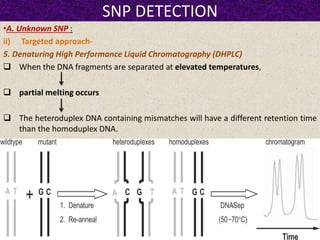
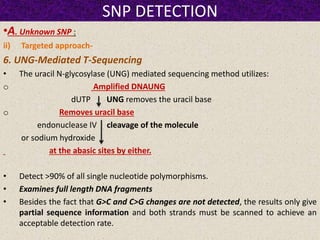
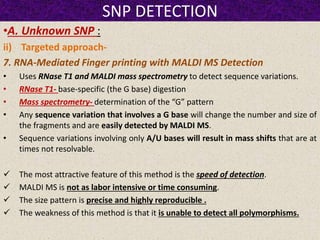
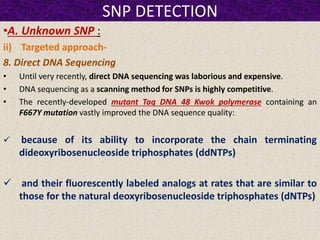
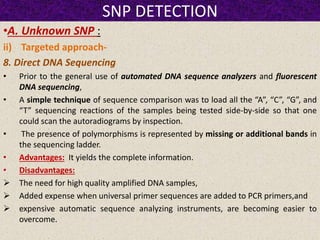
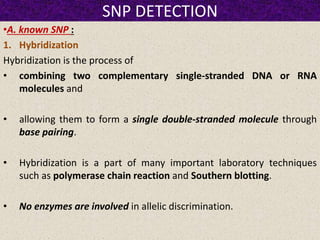
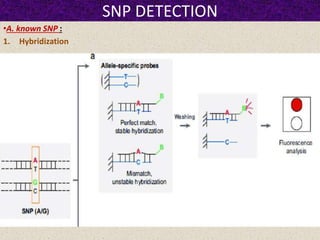
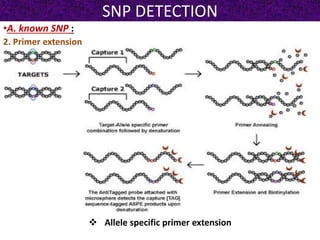
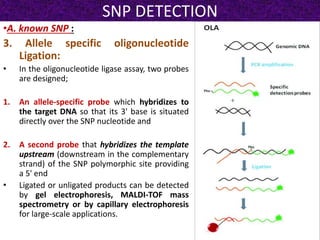
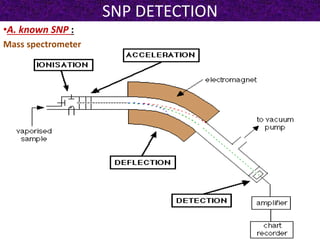
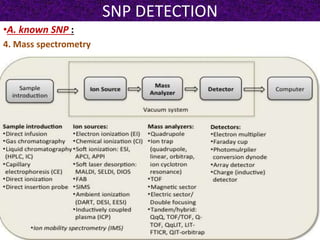
This document summarizes methods for detecting single nucleotide polymorphisms (SNPs). It discusses two main approaches: detecting unknown SNPs and detecting known SNPs. For unknown SNPs, global methods like restriction fragment length polymorphisms and targeted methods like denaturing gradient gel electrophoresis are used to scan DNA for new polymorphisms. For known SNPs, techniques like hybridization, primer extension, ligation, and mass spectrometry can be applied to screen individuals. Direct DNA sequencing is also discussed as the gold standard method, though historically it was labor-intensive. The document provides details on the principles and advantages/limitations of various SNP detection techniques.

![SNP
•SINGLE NUCLEOTIDE POLYMORPHISM
•The most common type of genetic variation among people.
•Each SNP represents a difference in a single DNA building block, called a nucleotides.
•DNA sequence variation occurring when a single nucleotide adenine (A), thymine (T),
cytosine (C), or guanine (G]) in the genome (or other shared sequence) differs
between members of a species or paired chromosomes in an individual.
•A minor allele frequency.
•Act as genetic markers/ molecular markers.](https://image.slidesharecdn.com/pptsnpdetection-201028150535/85/Ppt-snp-detection-3-320.jpg)