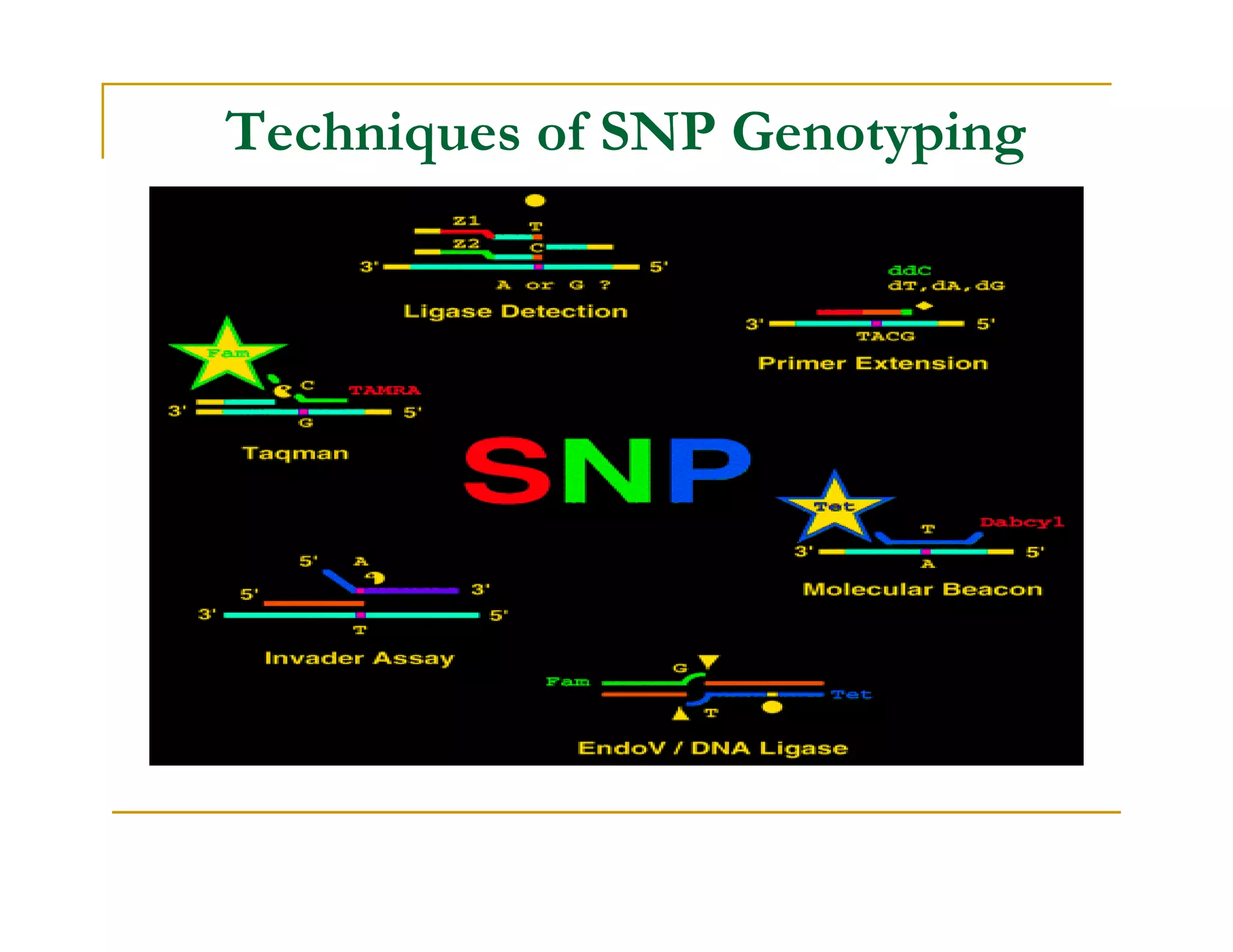
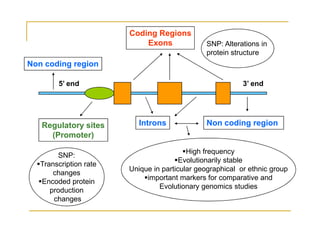
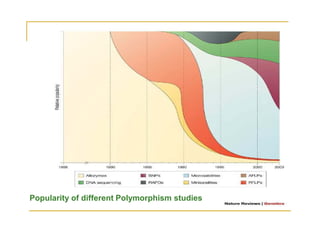
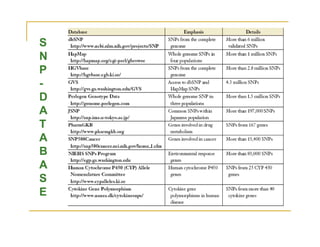
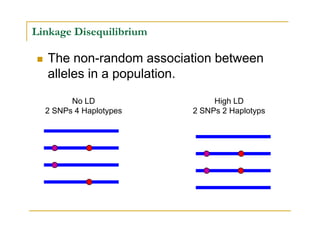
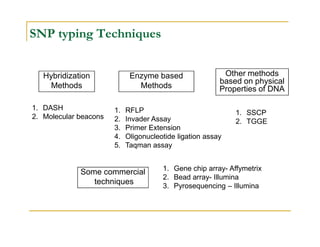
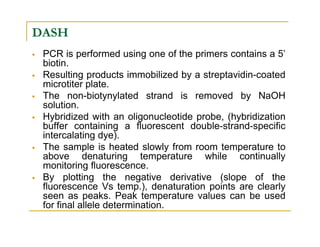
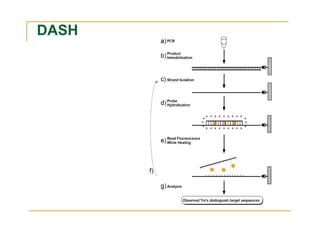
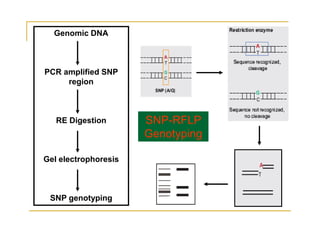
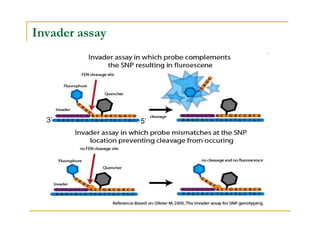
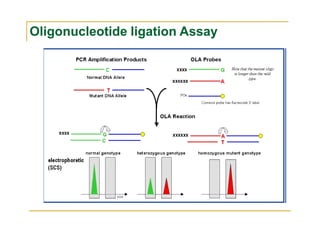
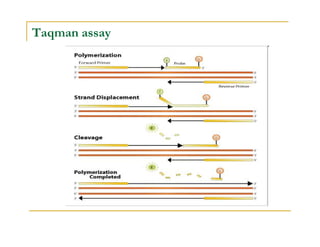
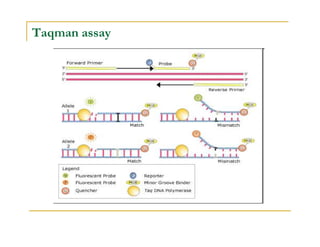
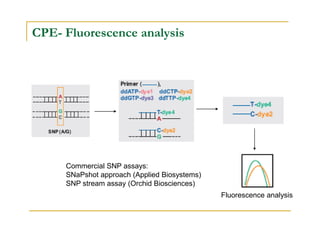
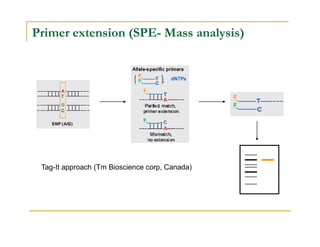
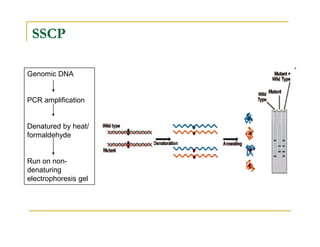
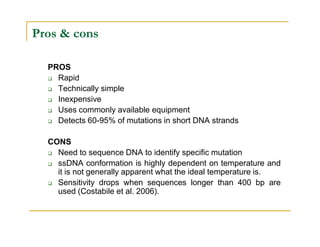
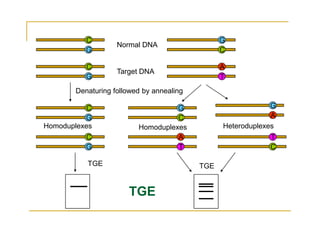
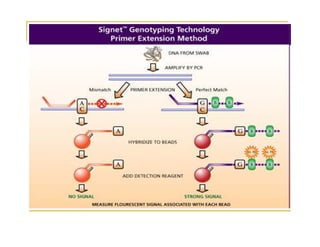
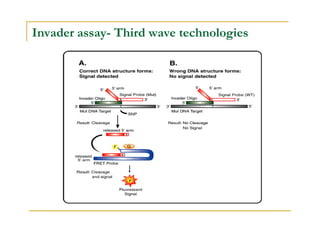
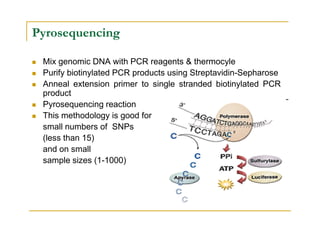
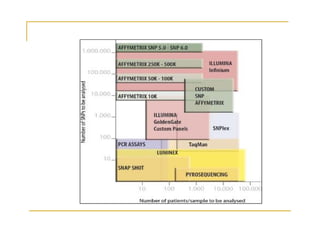
Techniques of SNP Genotyping can be summarized as follows:
There are several techniques for genotyping SNPs including hybridization methods, enzyme-based methods, and other methods based on physical properties of DNA. Popular hybridization methods include DASH, molecular beacons, and gene chip arrays. Common enzyme-based techniques are RFLP, Invader assay, and oligonucleotide ligation assay. Other physical property-based methods include SSCP, TGGE, and pyrosequencing. Each method has its own pros and cons related to factors like speed, cost, and accuracy. Choosing the appropriate SNP genotyping technique depends on the number of SNPs needed to be analyzed and sample size.