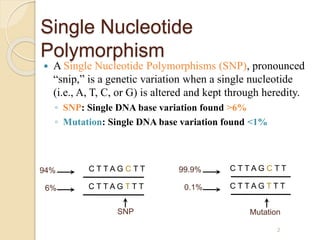
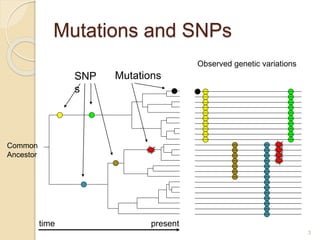
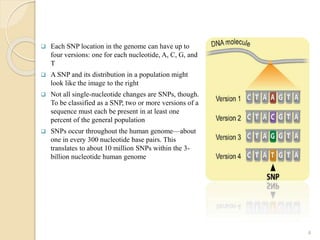
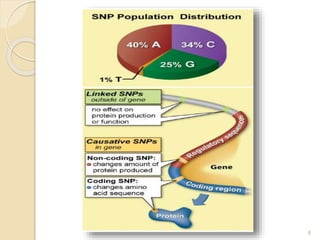
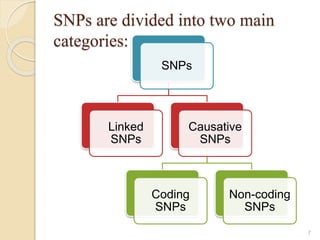
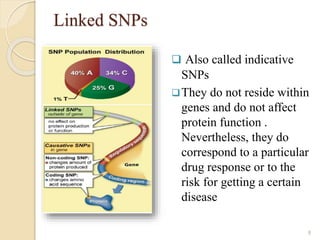
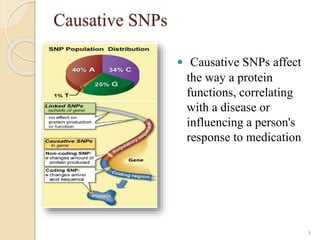
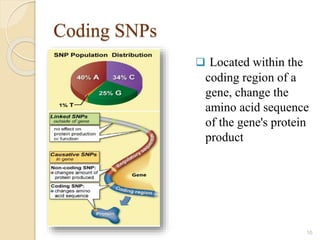
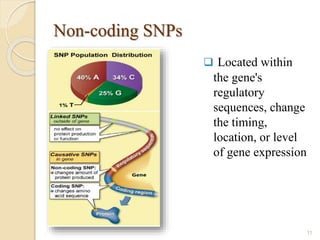
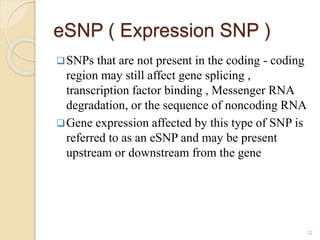
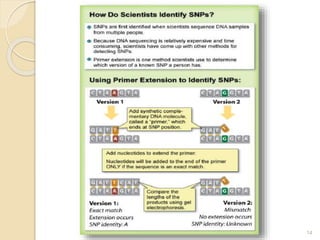
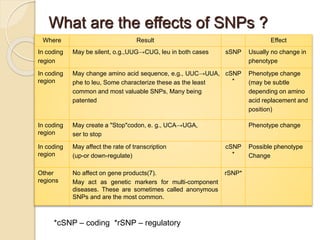
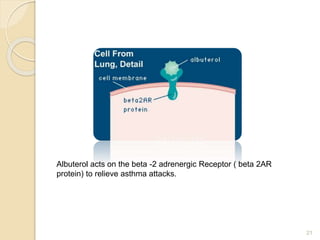
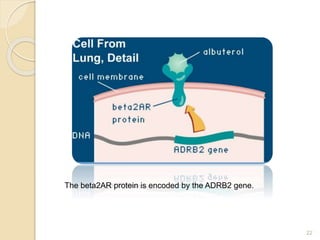
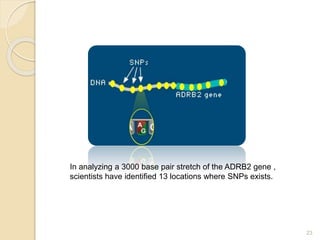
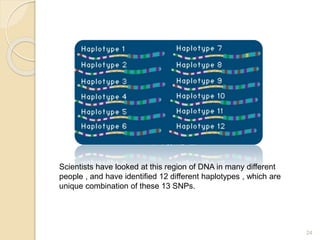
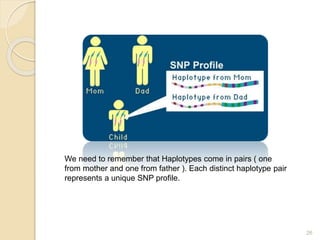
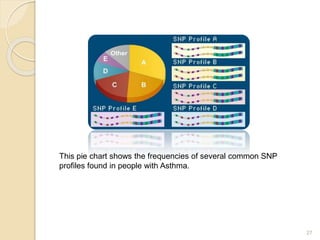
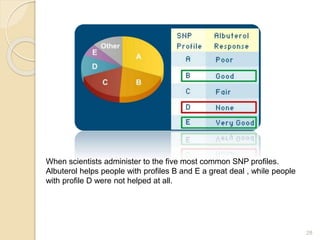
The document discusses single nucleotide polymorphisms (SNPs), which are genetic variations impacting human genomes. It differentiates between SNPs and mutations, outlines their classification into linked and causative SNPs, and emphasizes their potential effects on disease and drug responses. Additionally, it highlights ongoing research into how SNP profiles can influence treatments like albuterol for asthma.