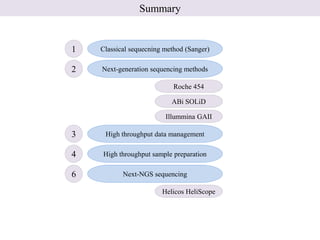
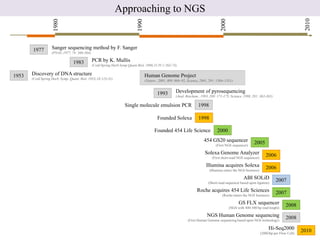
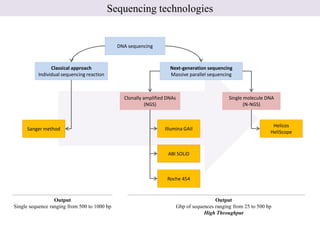
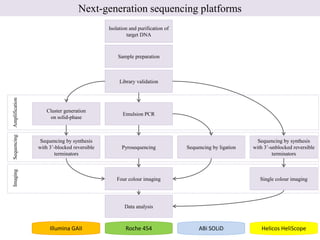
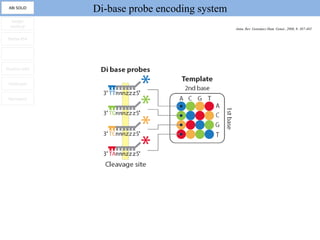
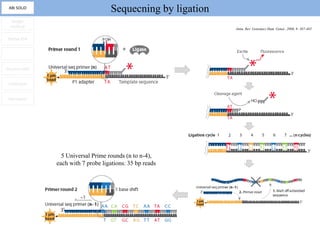
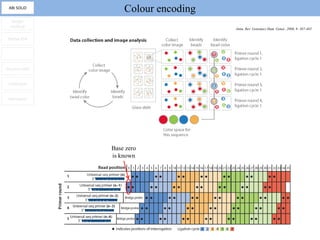
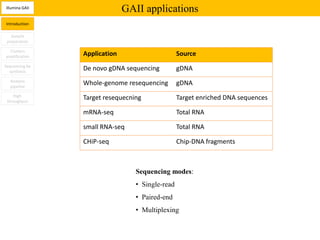
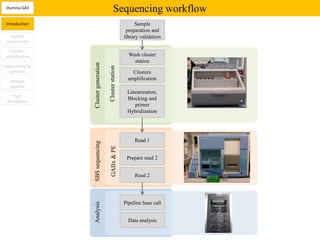
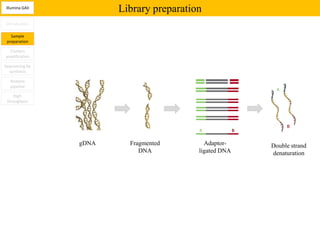
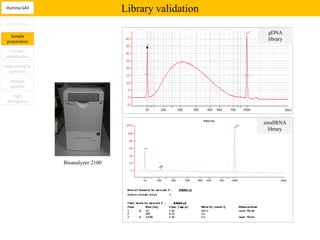
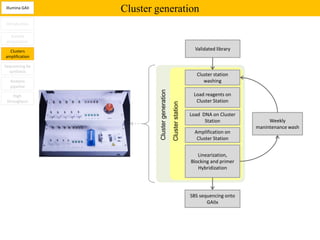
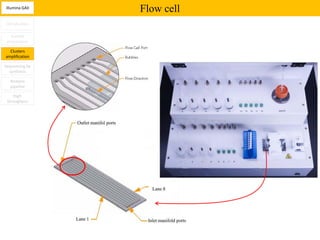
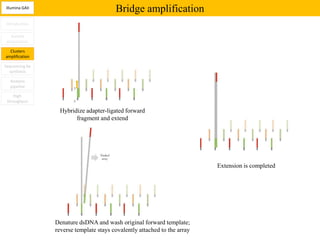
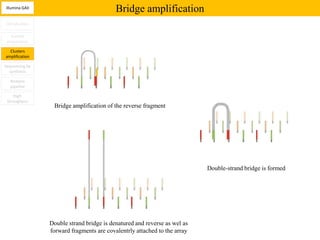
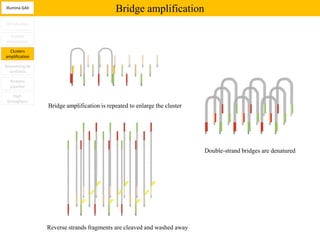
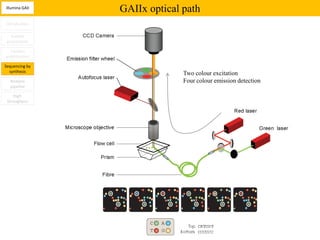
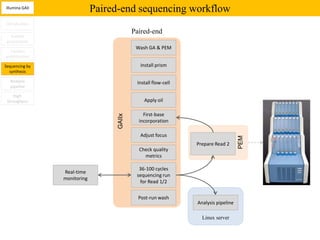
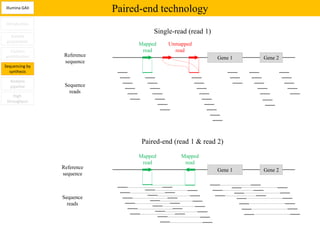
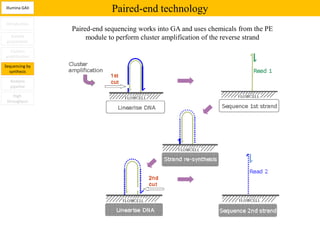
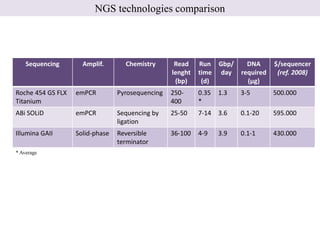
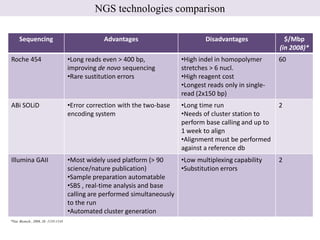
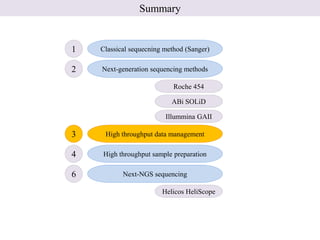
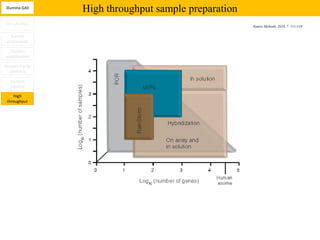
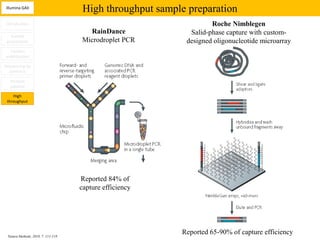
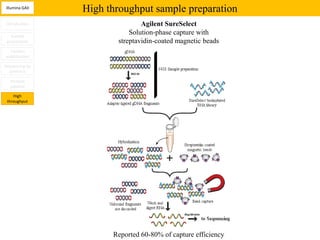
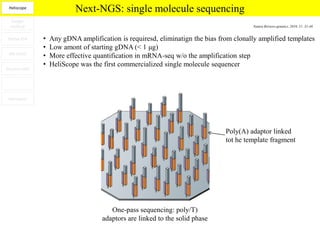
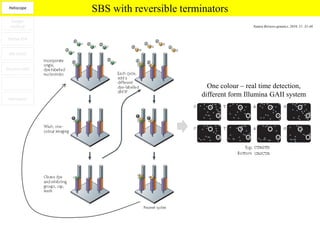
1. Next-generation sequencing methods such as Roche 454, Illumina GAII, and ABI SOLiD allow for high throughput DNA sequencing through massive parallel sequencing.
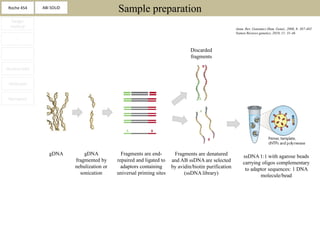
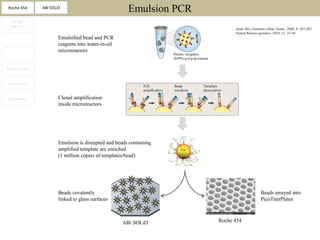
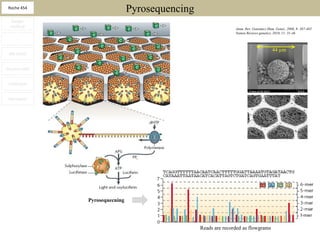
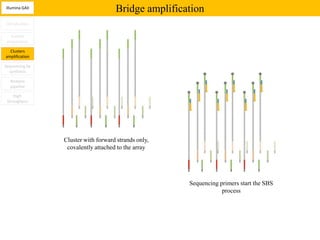
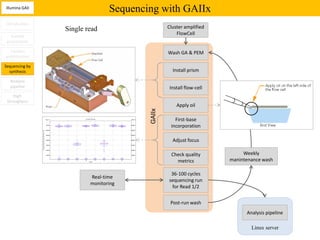
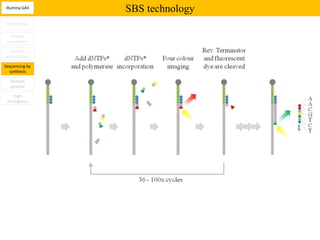
2. These methods involve clonal amplification of DNA fragments on solid surfaces or in emulsion PCR followed by sequencing using pyrosequencing, sequencing by synthesis with reversible terminators, or sequencing by ligation approaches.
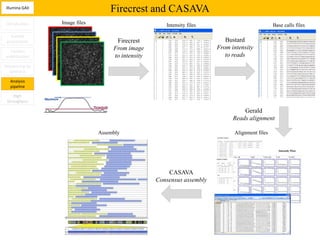
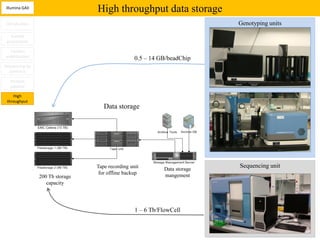
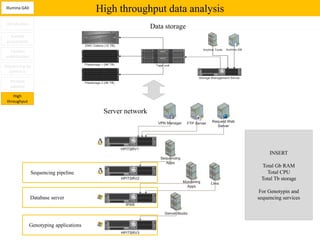
3. The resulting sequencing data requires high throughput management and analysis pipelines to process the large volumes of sequence data produced.