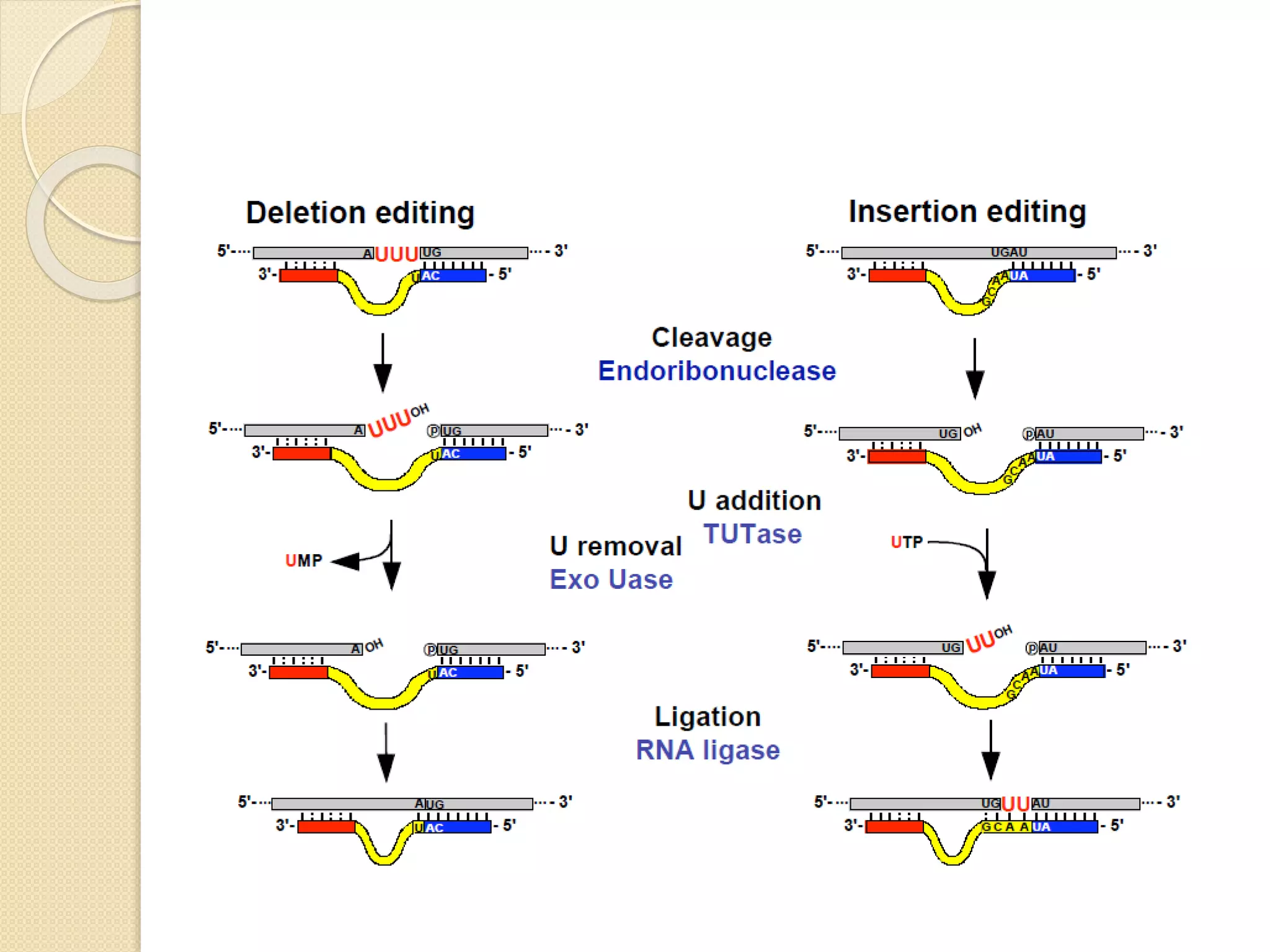
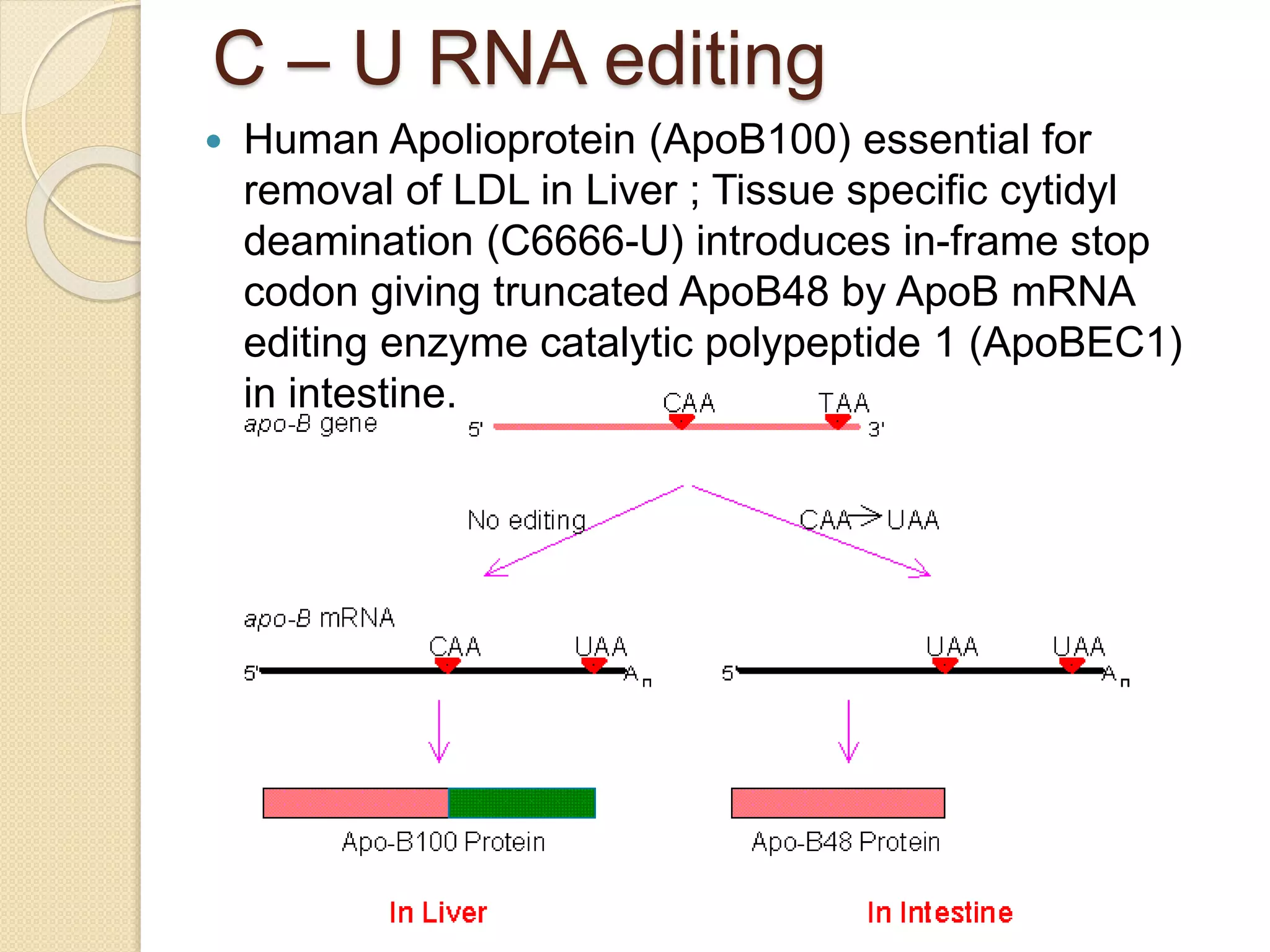
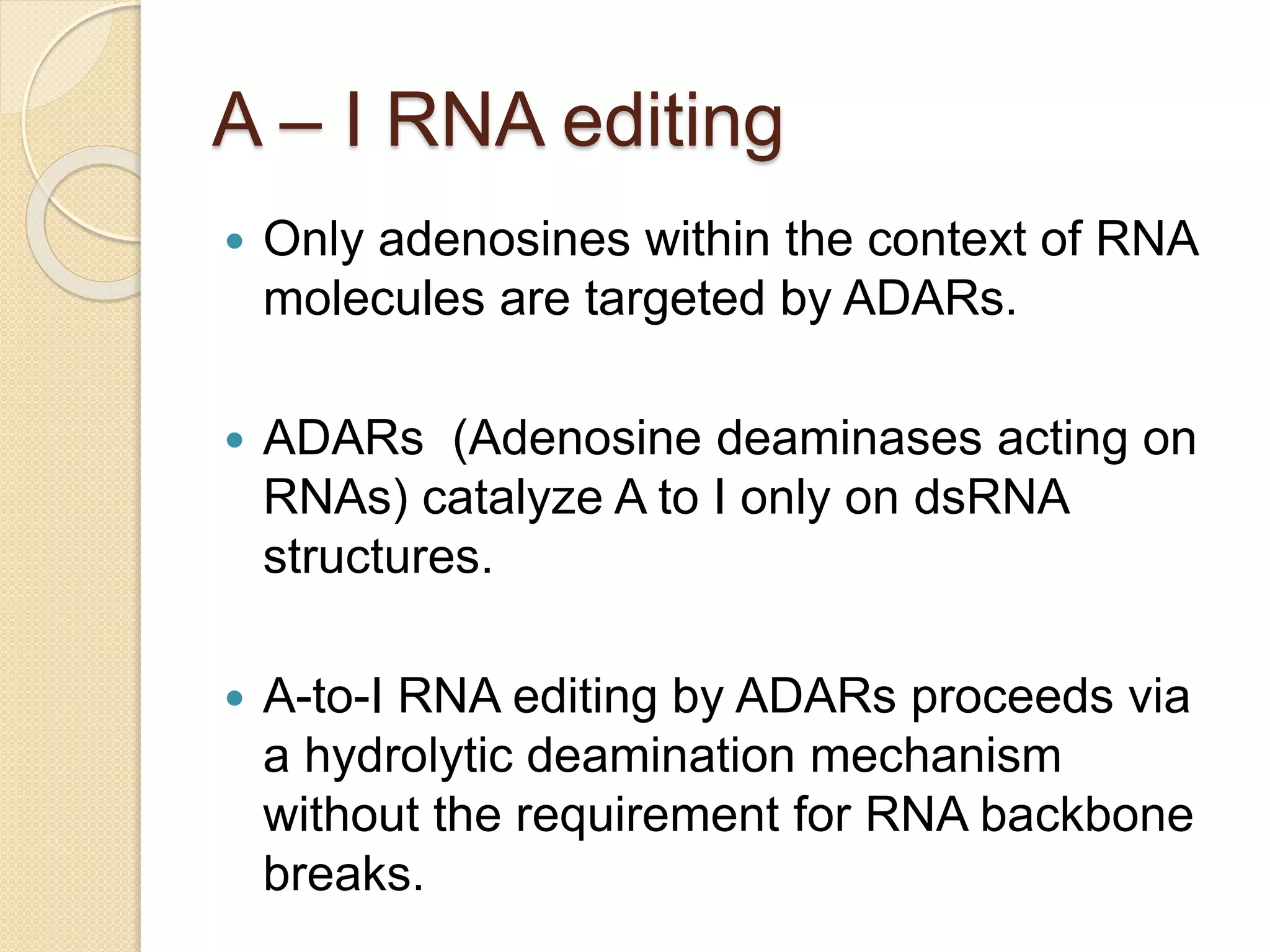
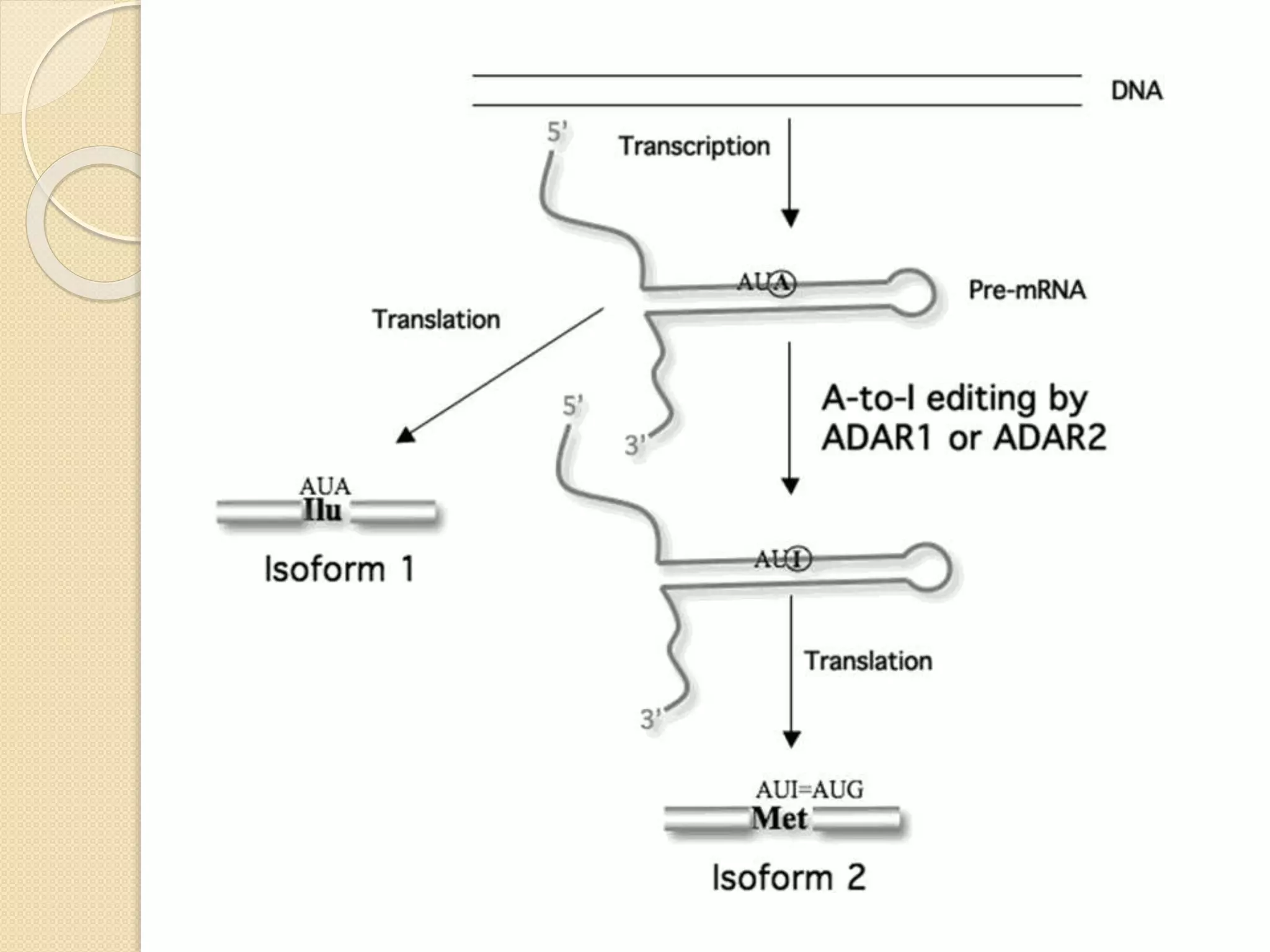
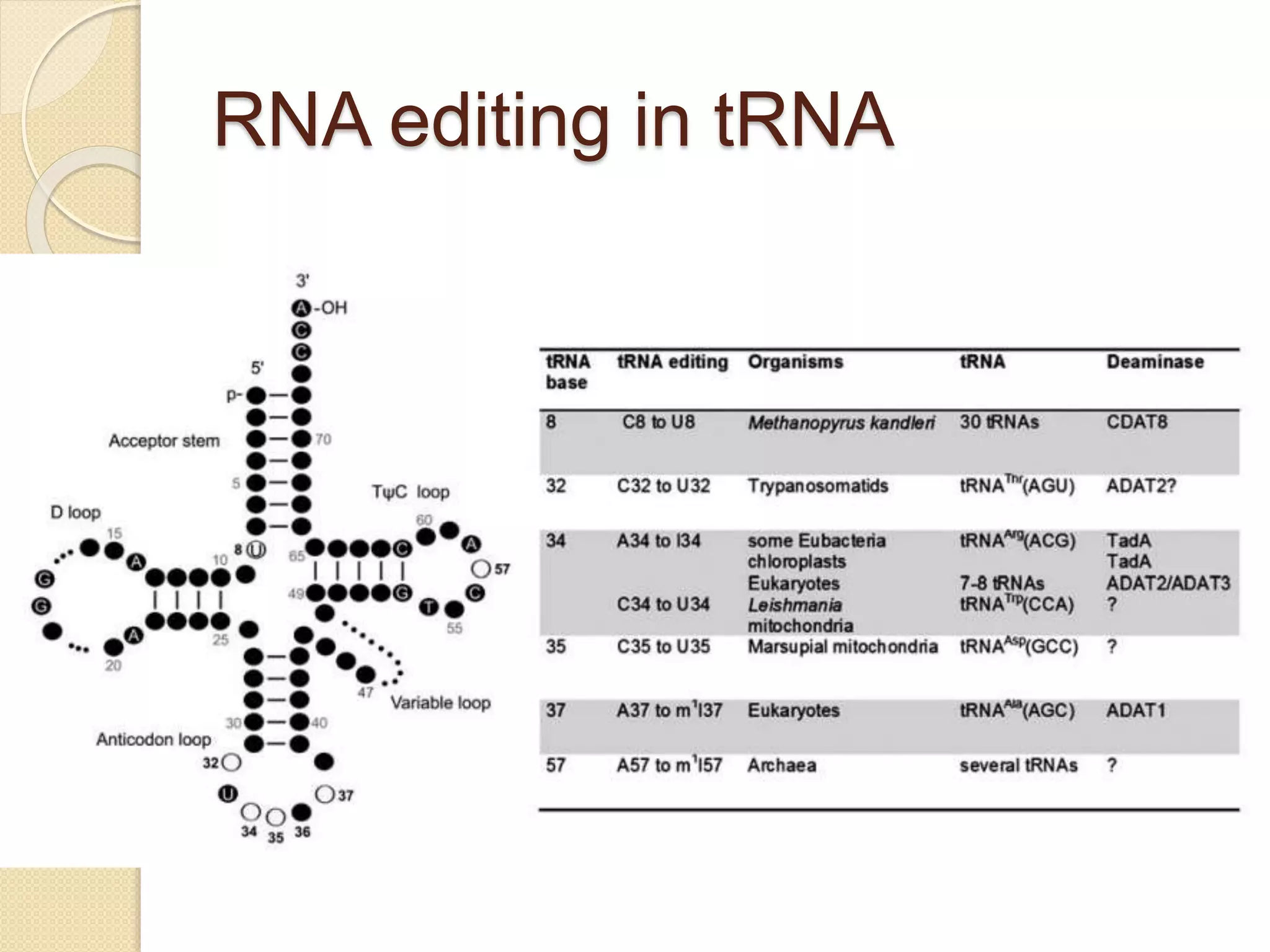
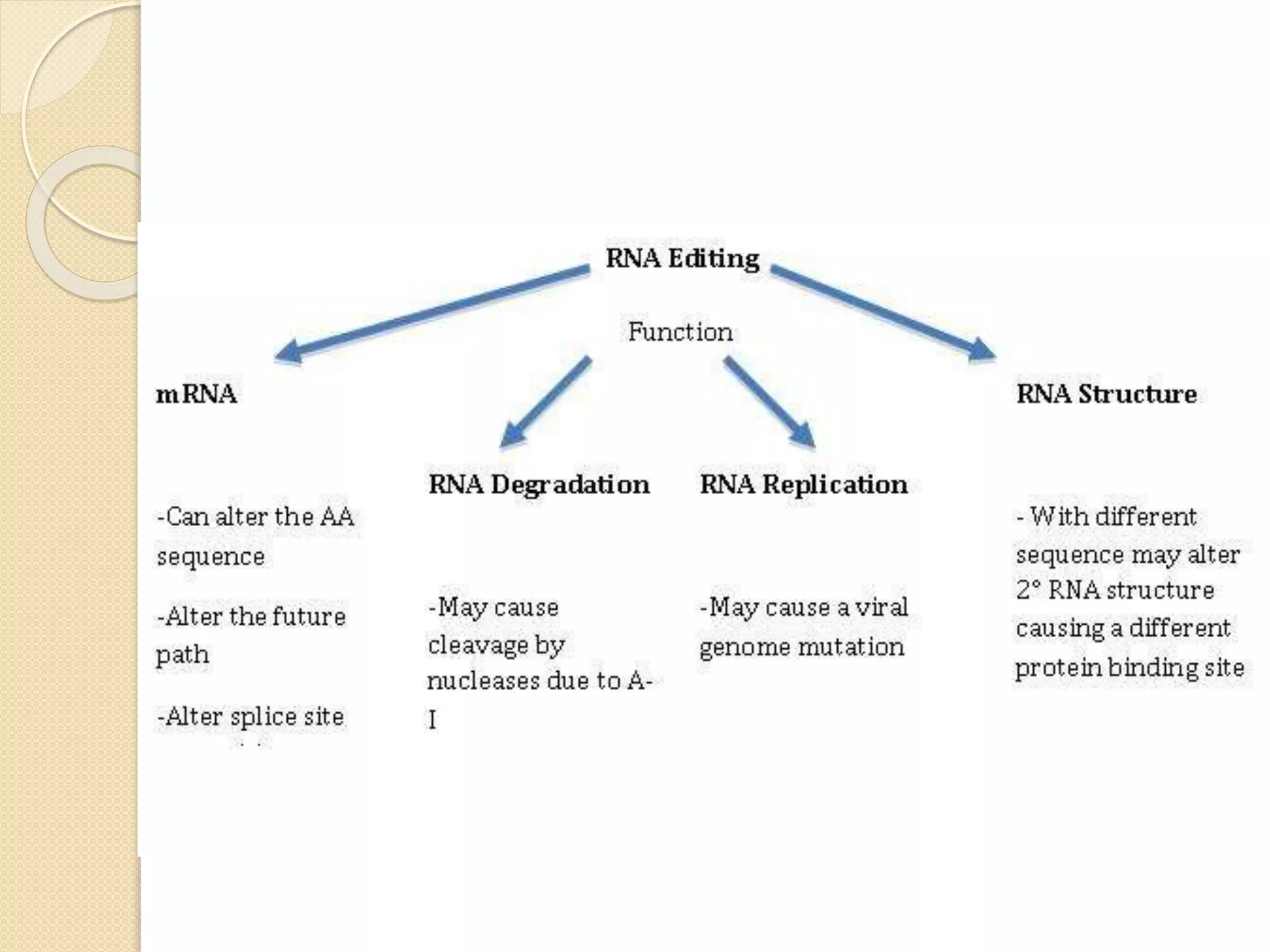
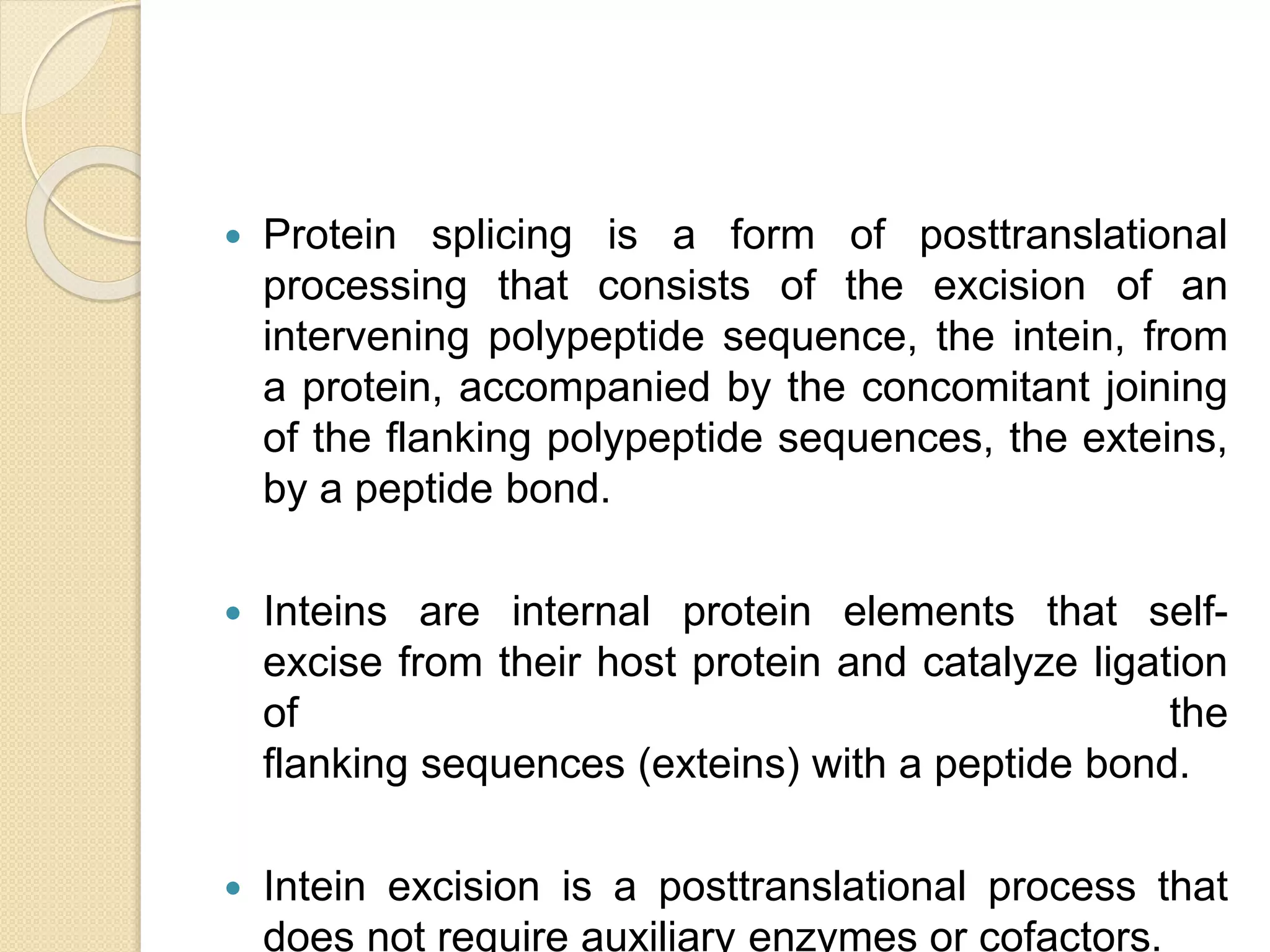
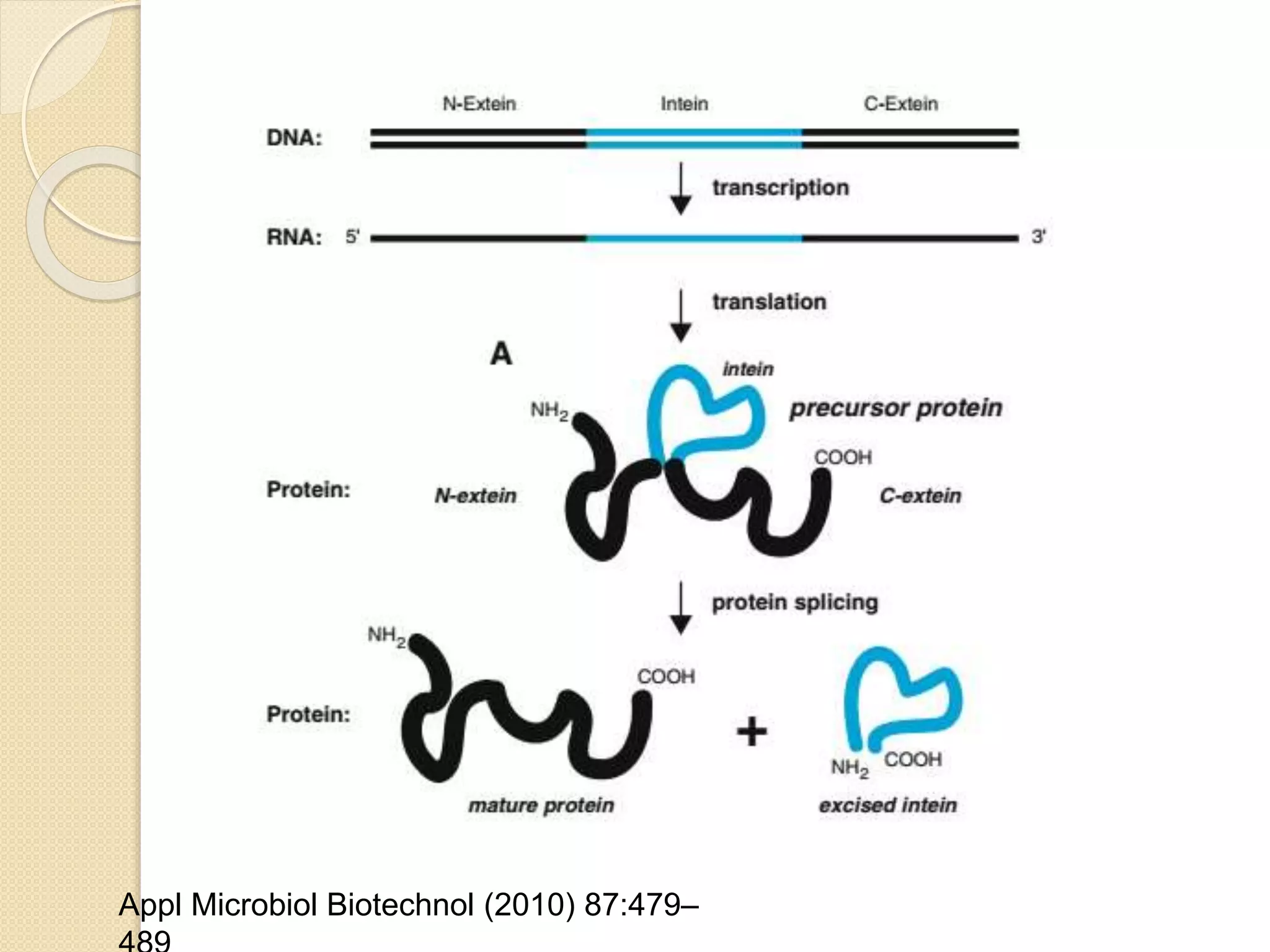
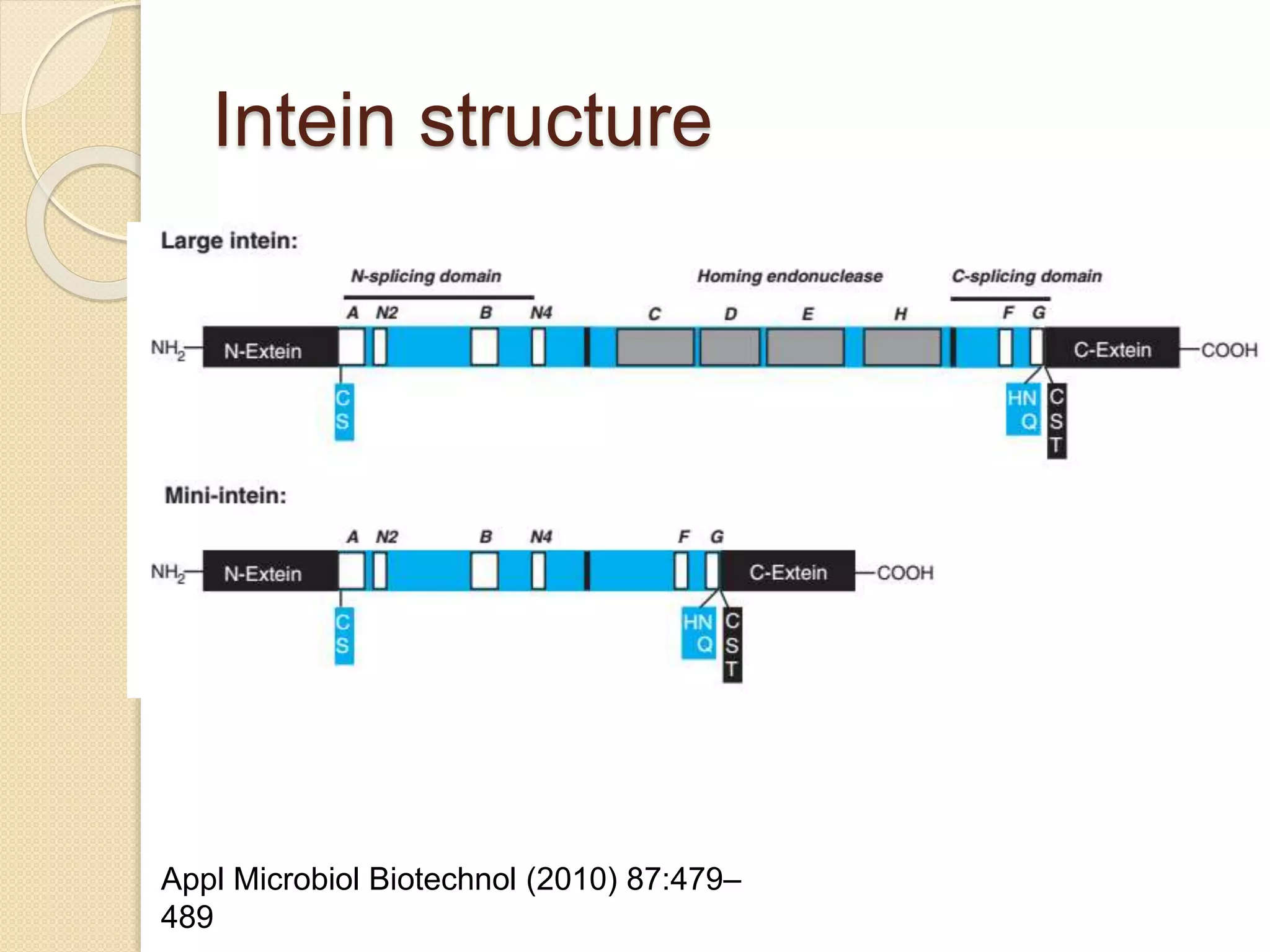
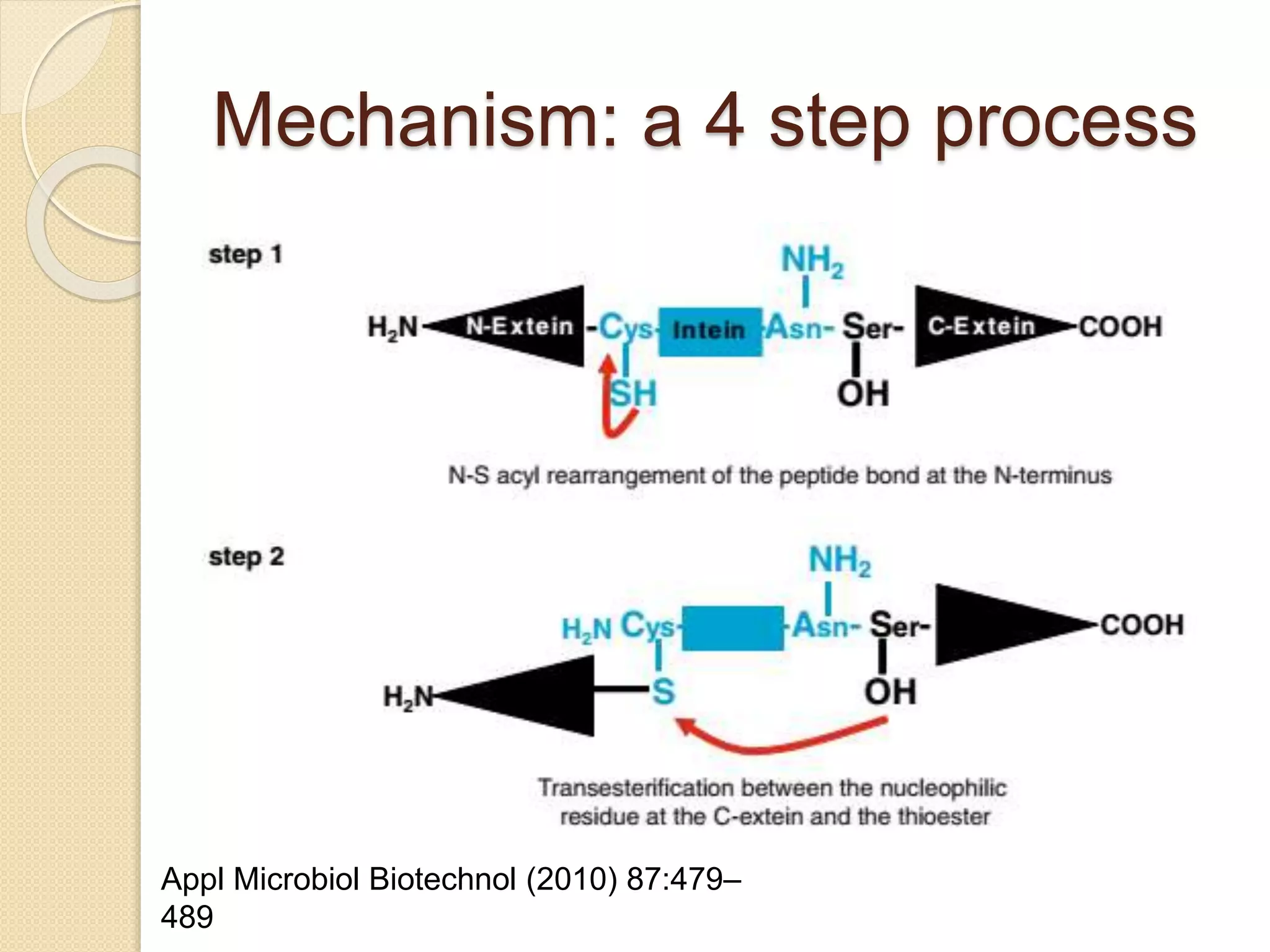
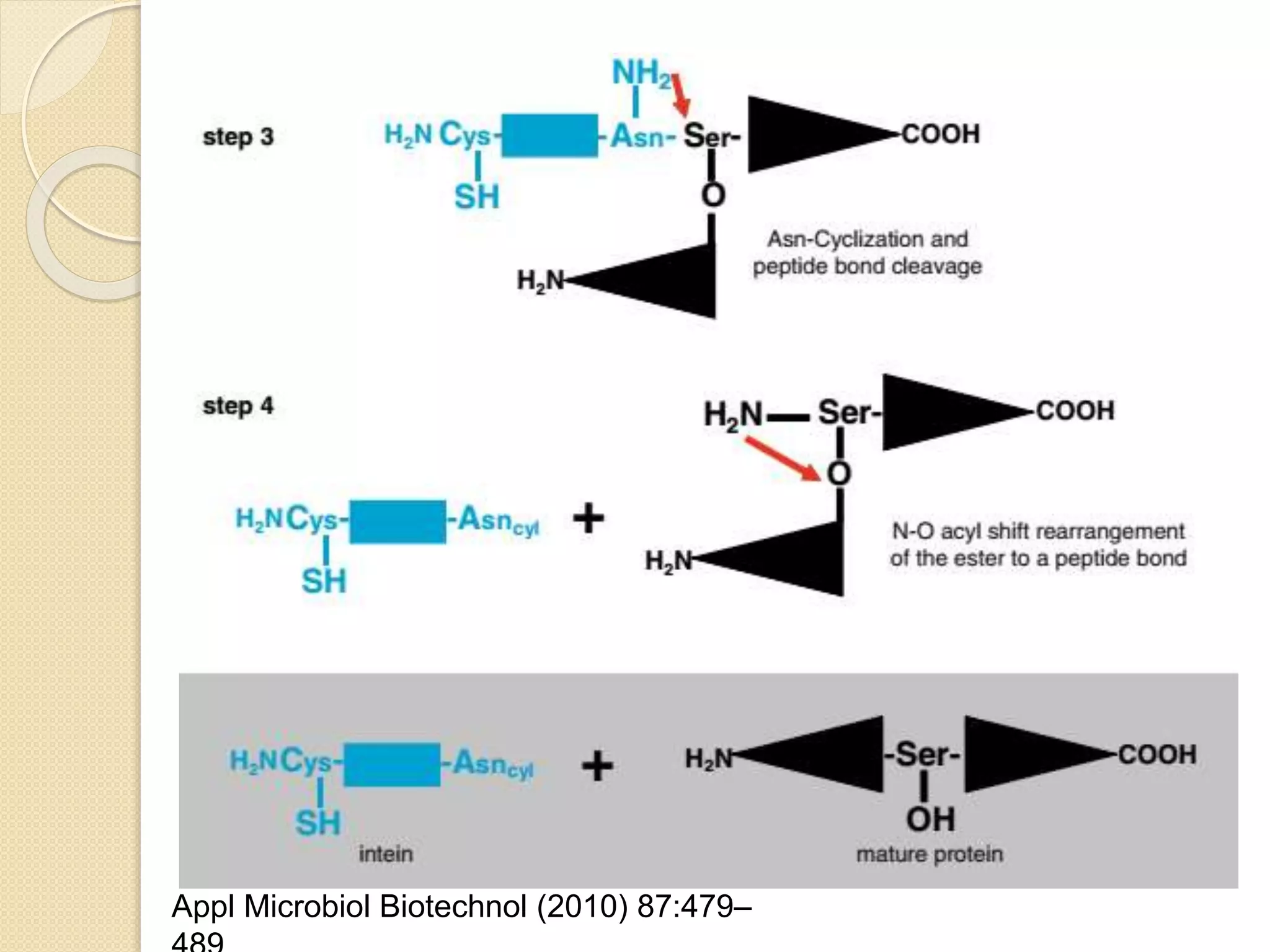
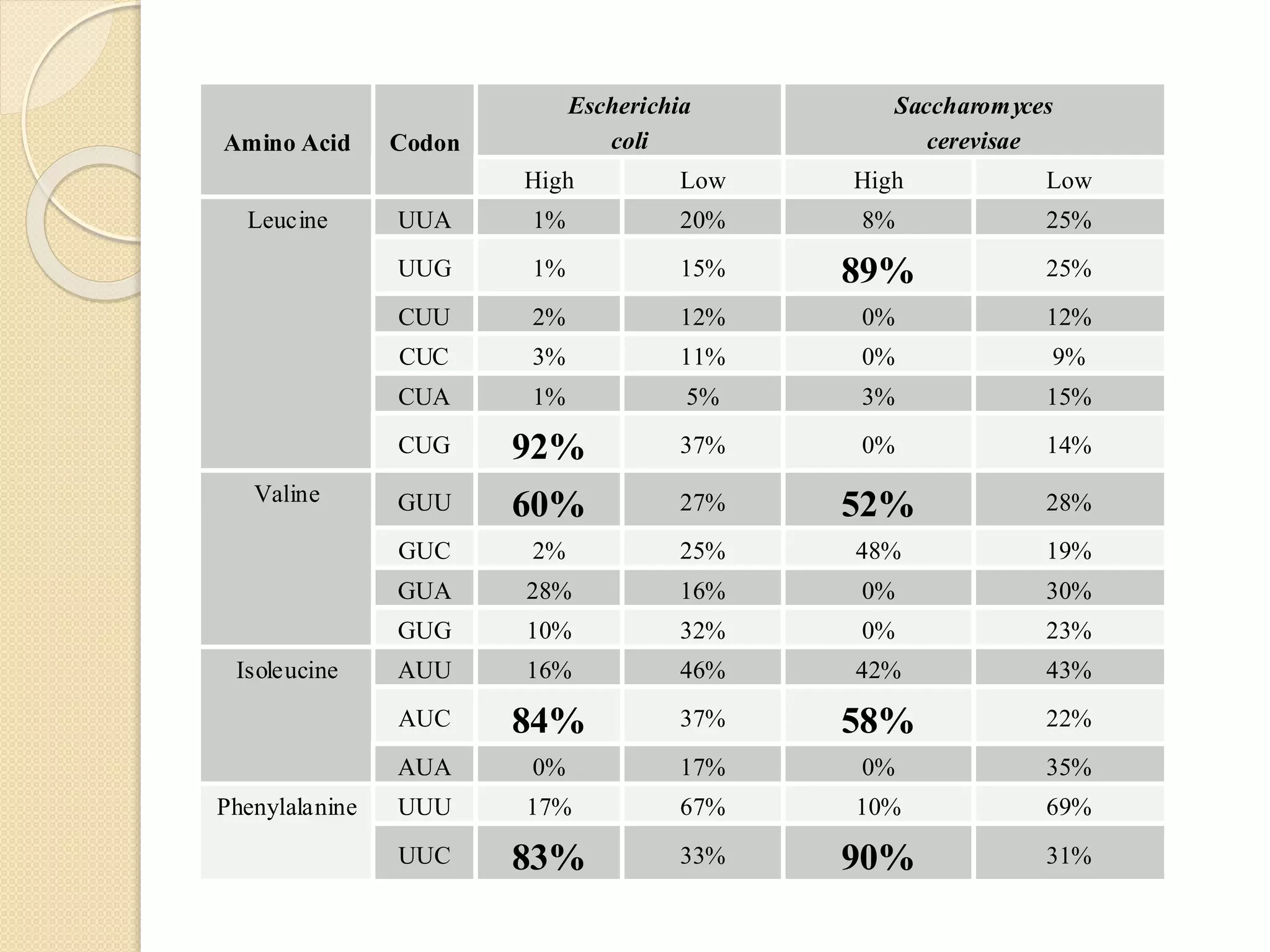
RNA editing is any process that alters the sequence of an RNA transcript from that of the DNA template. It was first discovered in trypanosome mitochondria but also occurs commonly in plant mitochondria and a few mammalian nuclear and chloroplast genes. RNA editing can produce multiple proteins from a single gene. Mechanisms include base modification by guide RNAs or enzymes that catalyze changes like A to I. Protein splicing is a posttranslational process where an intein protein sequence excises itself and ligates the flanking extein sequences. Codon bias refers to the preferential usage of certain codons over synonymous alternatives and can influence translation efficiency, accuracy and protein folding. Selection and mutation pressures contribute to differences in codon bias patterns between organisms