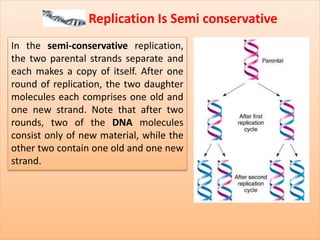
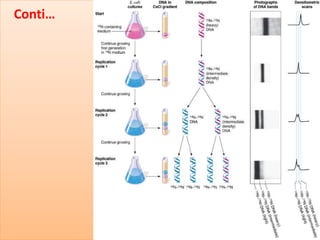
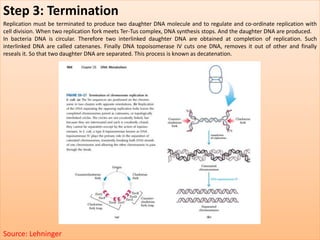
The document discusses DNA replication, detailing its essential role in cell division and the enzymes involved in the process. It describes key steps, including initiation, elongation, and termination, as well as the semi-conservative nature of replication illustrated by the Meselson & Stahl experiment. Additionally, specific enzymes like primase, DNA polymerase, helicase, ligase, and others are outlined for their functions in DNA replication.