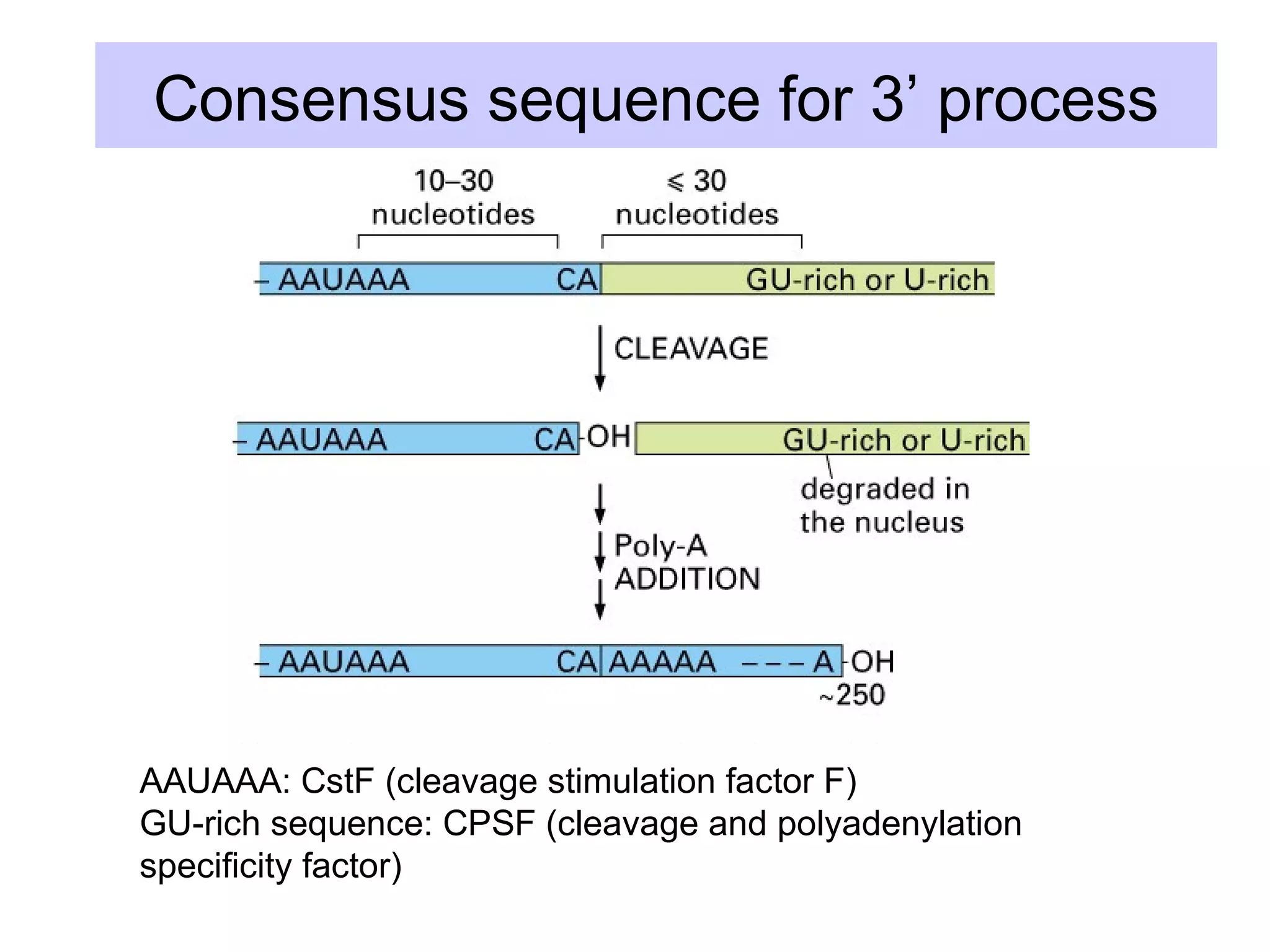
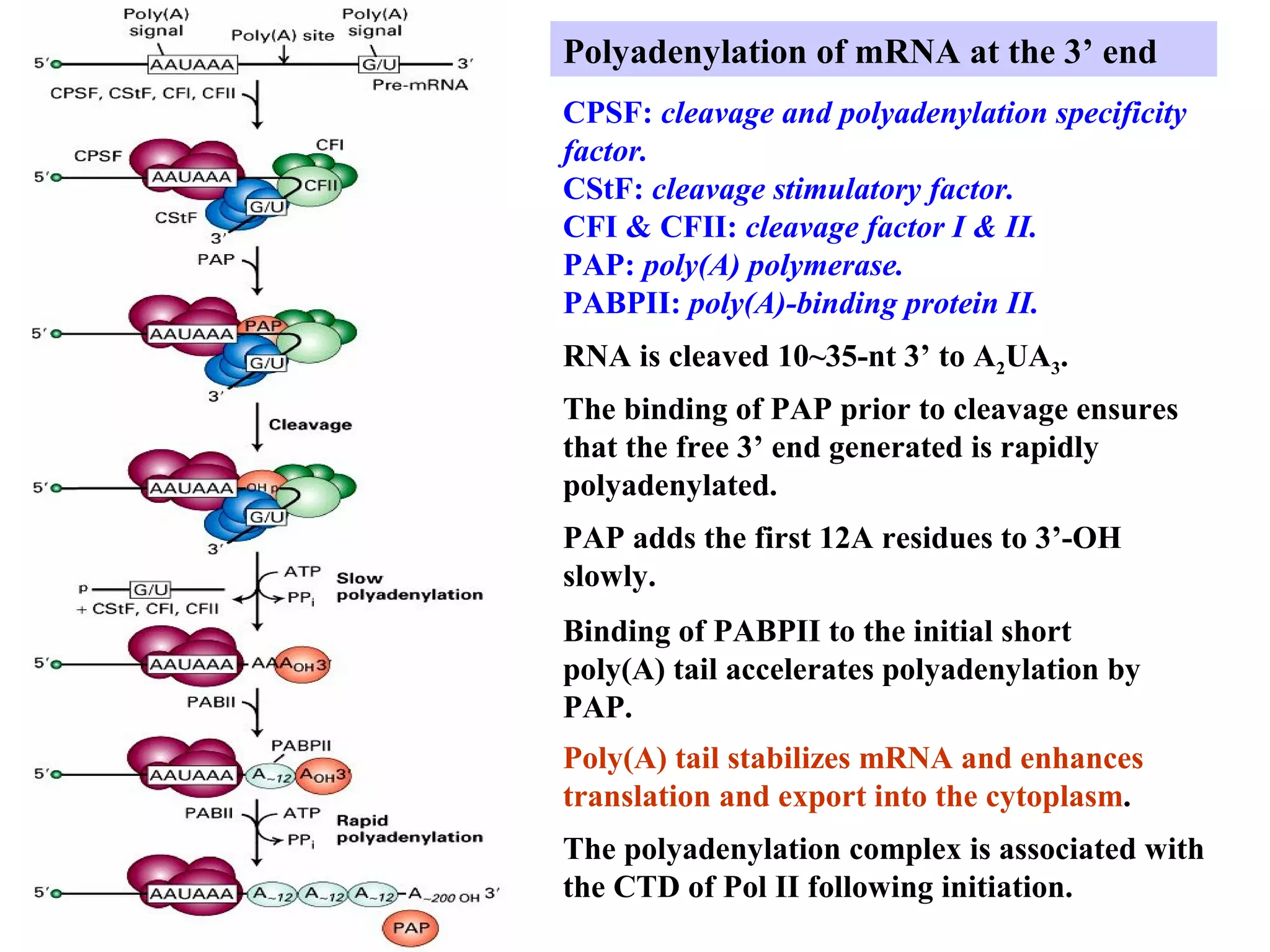
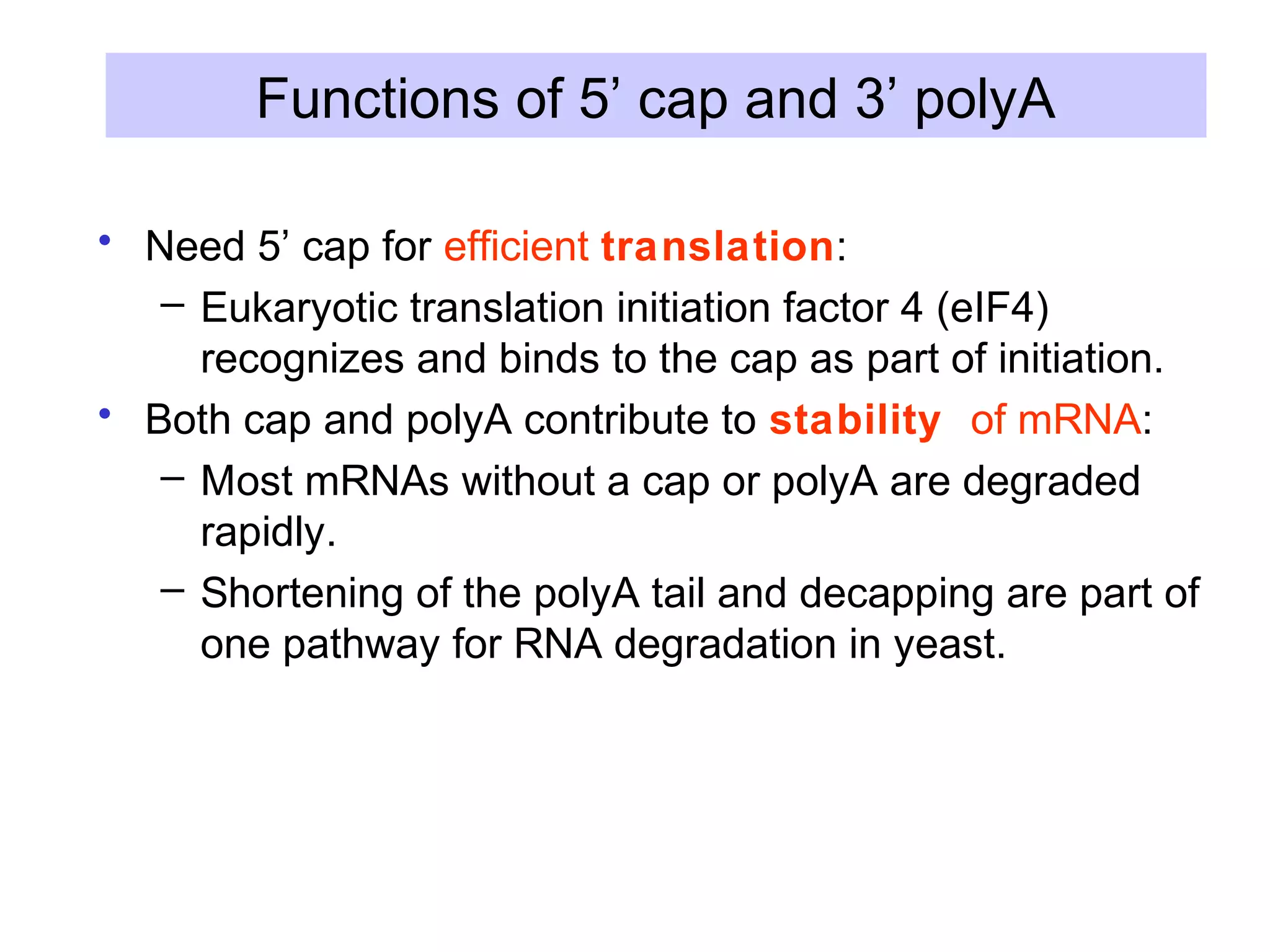
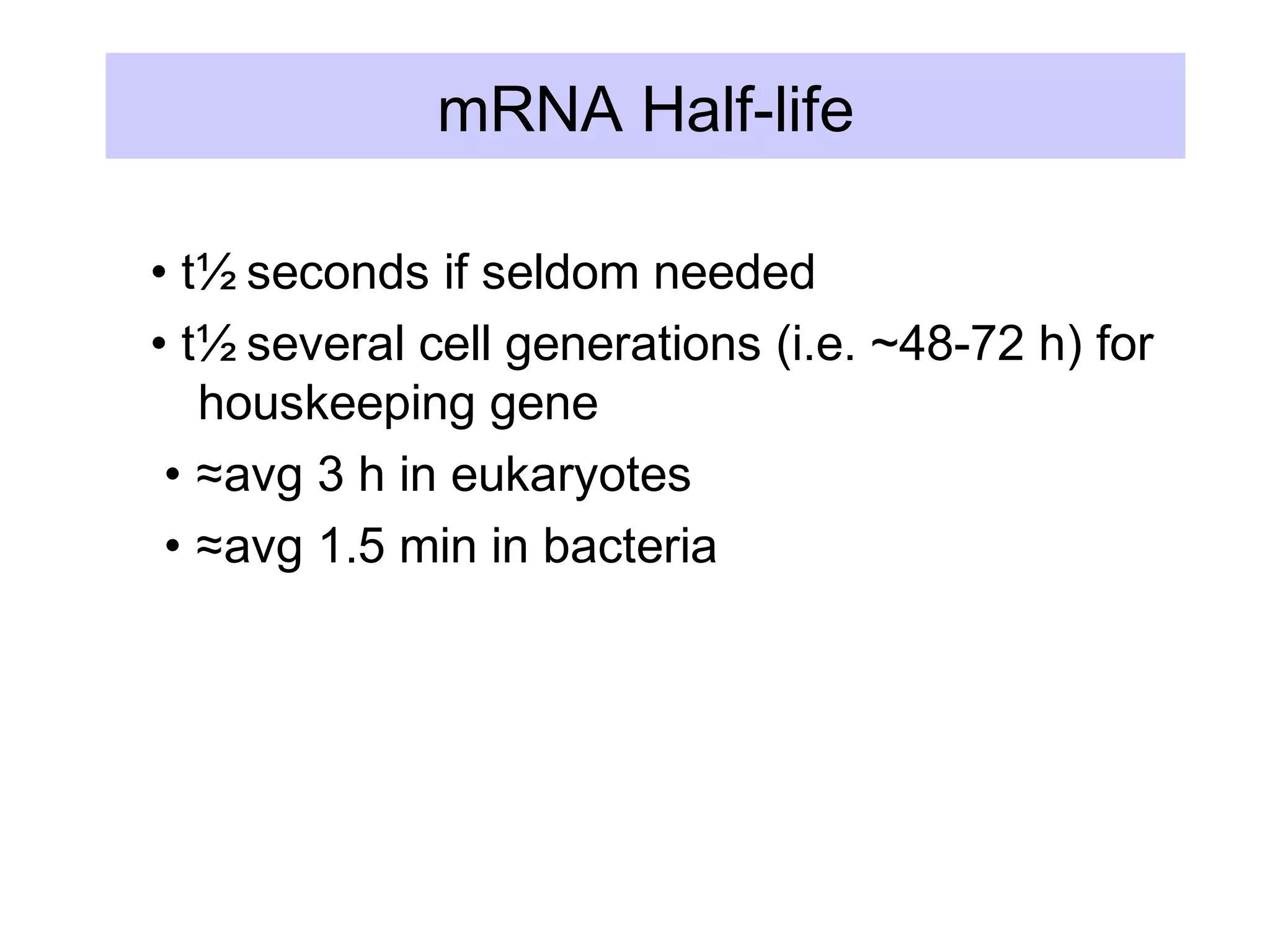
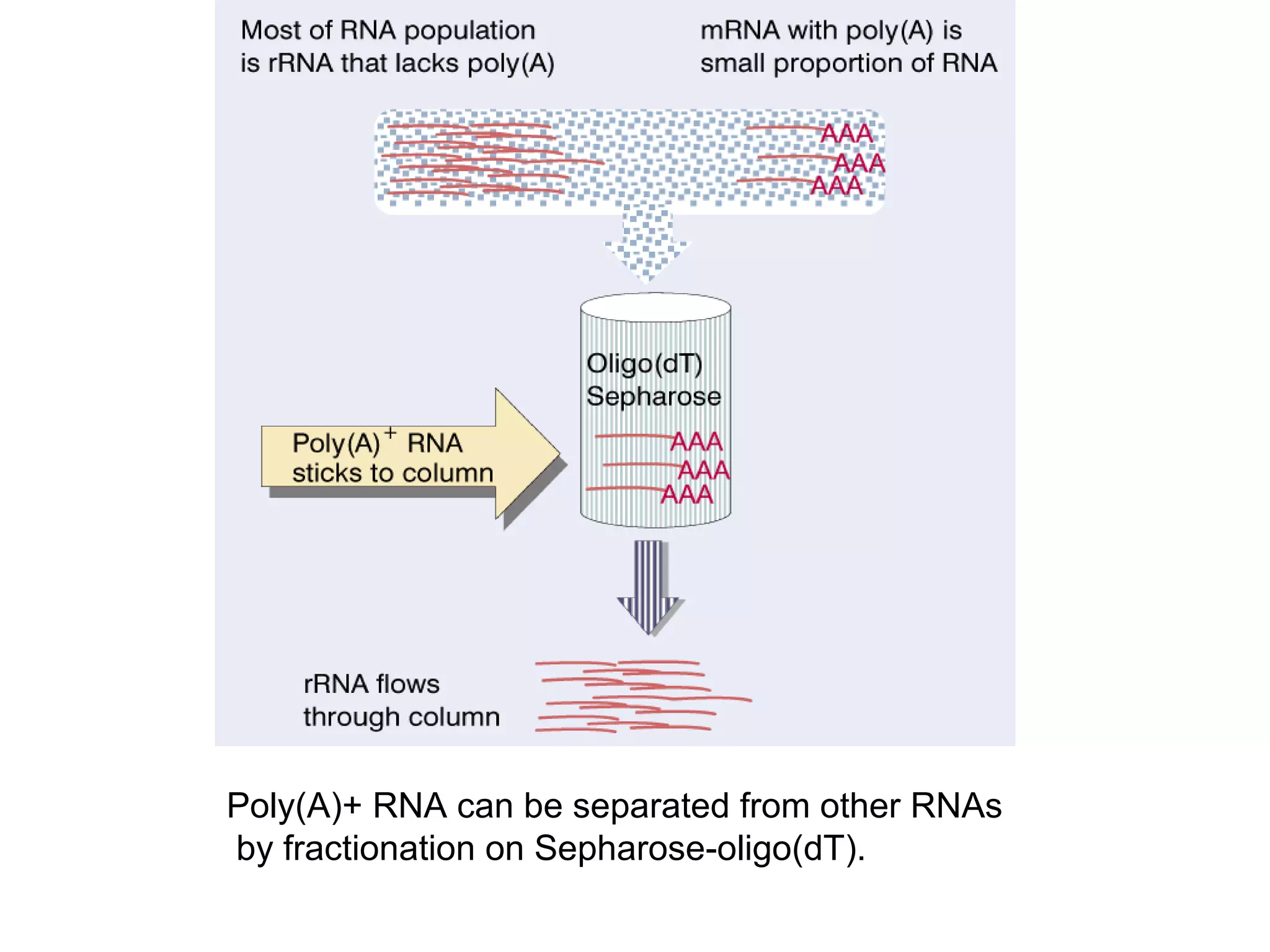
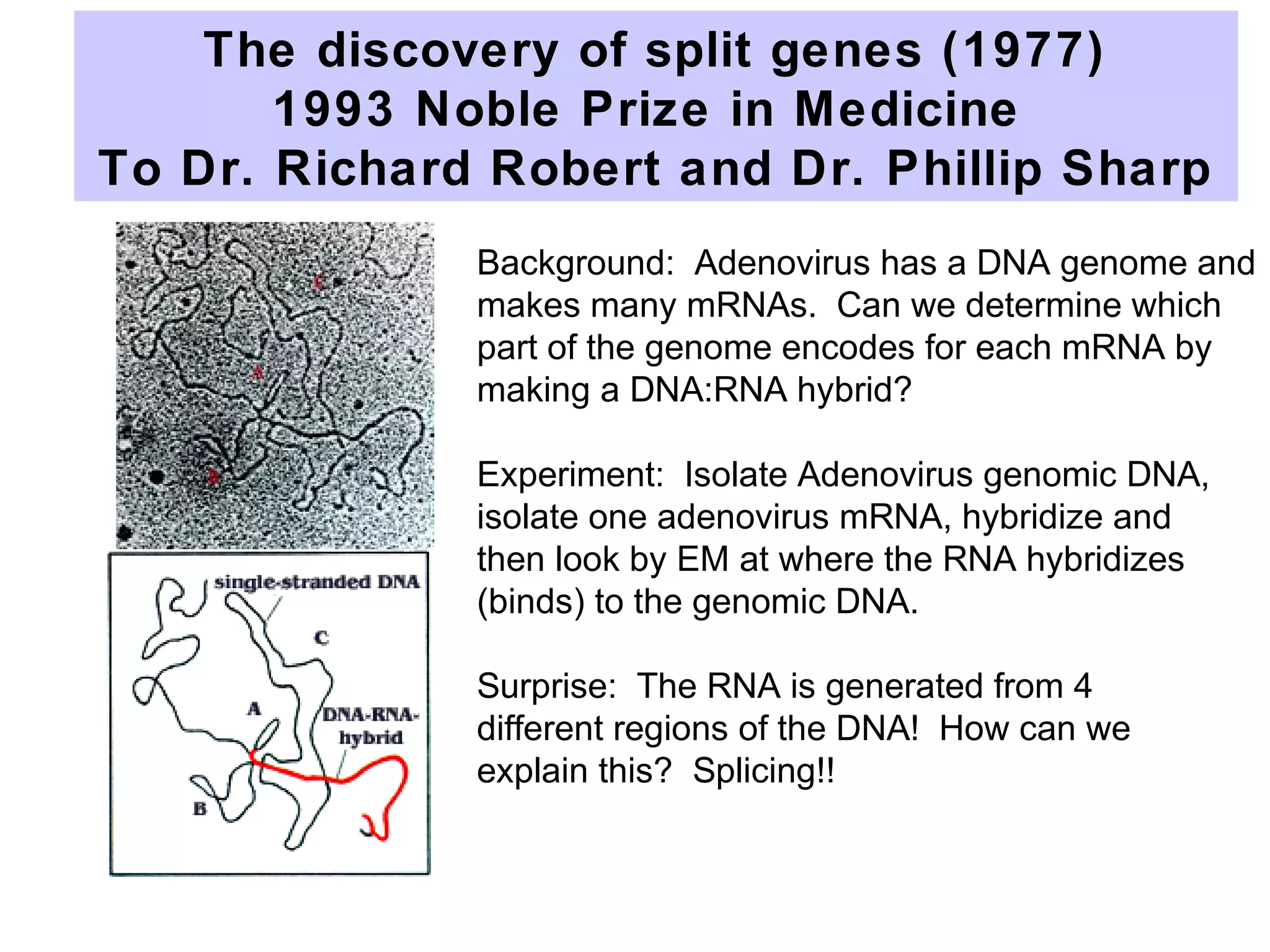
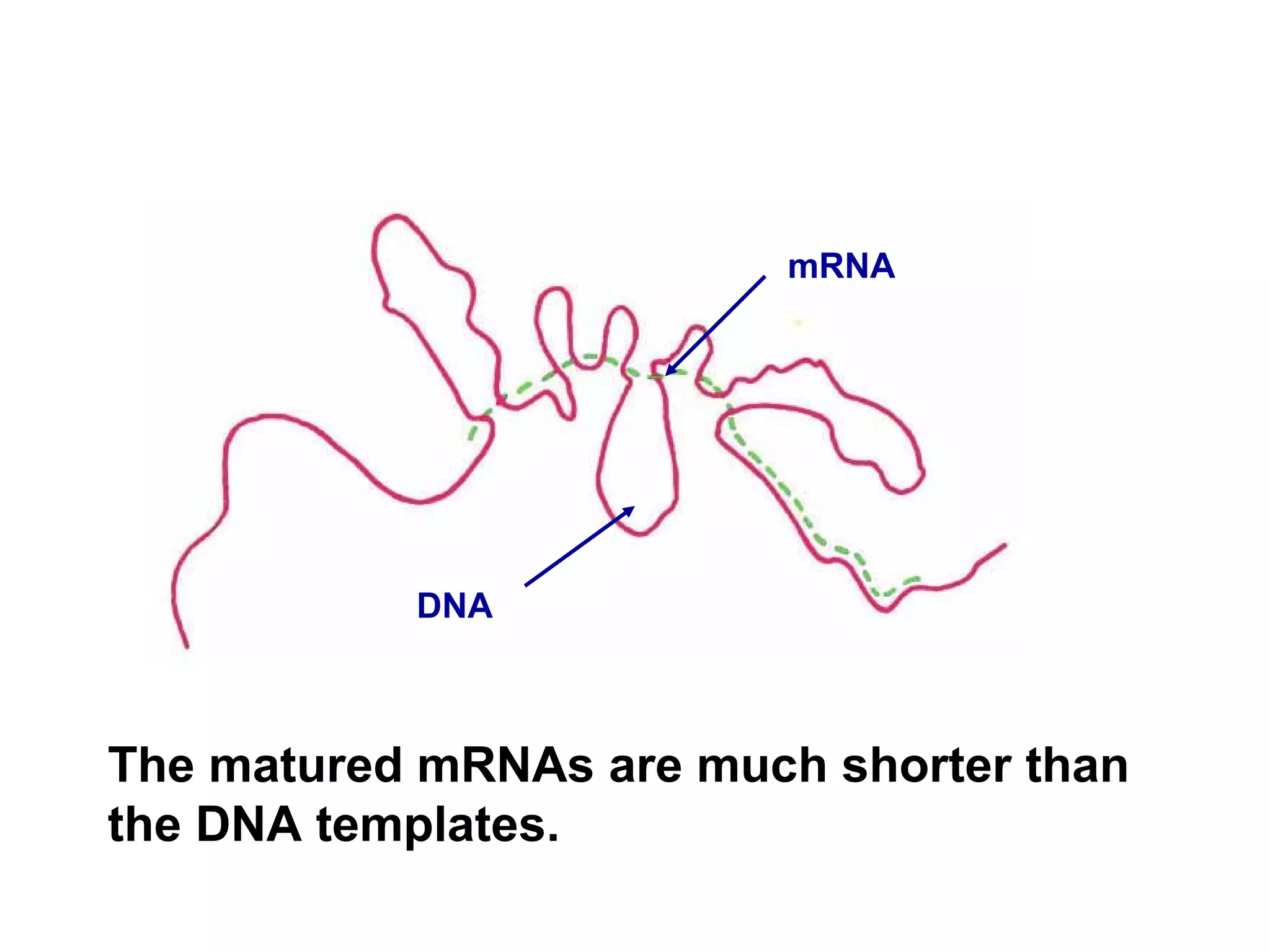
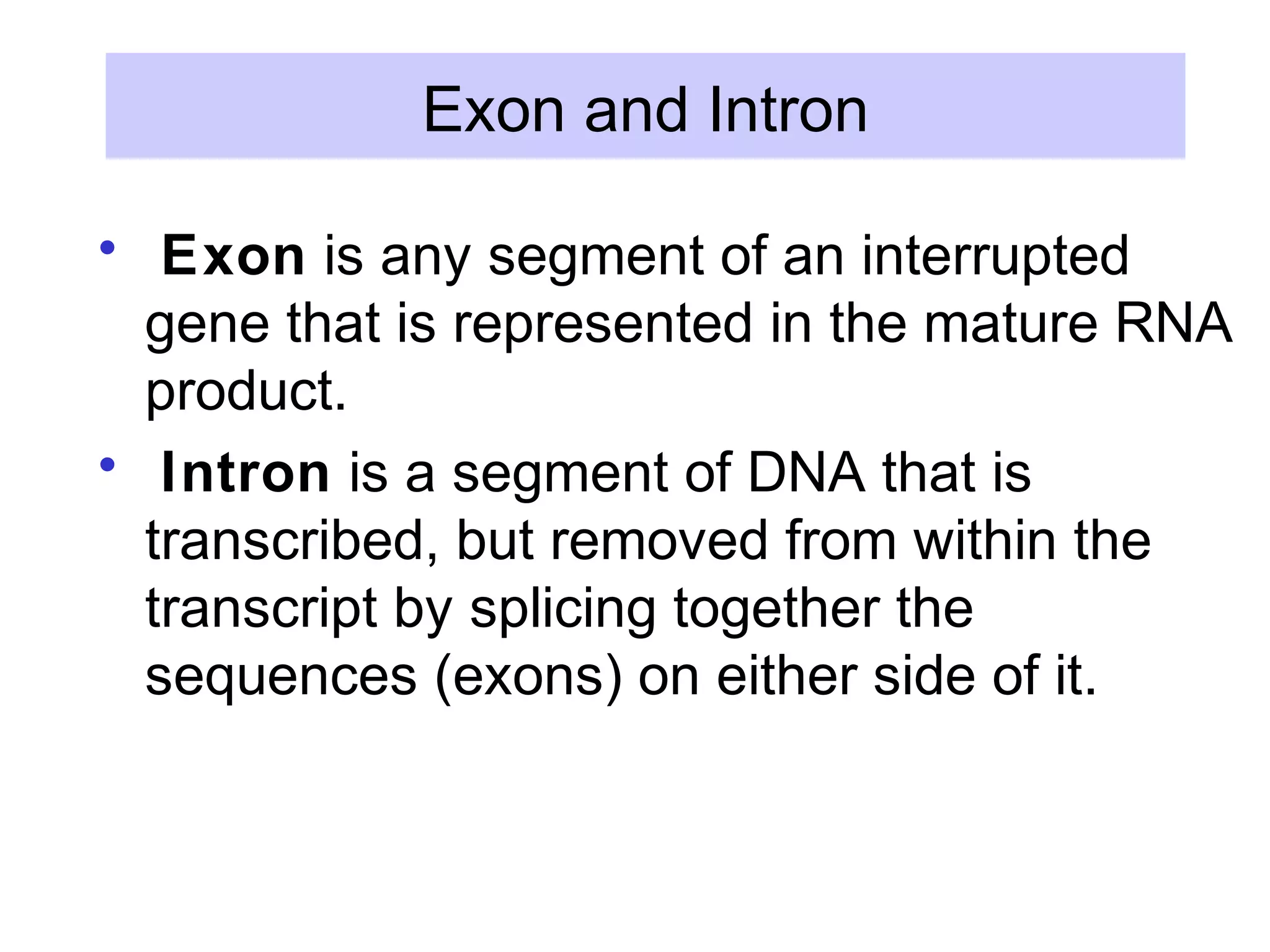
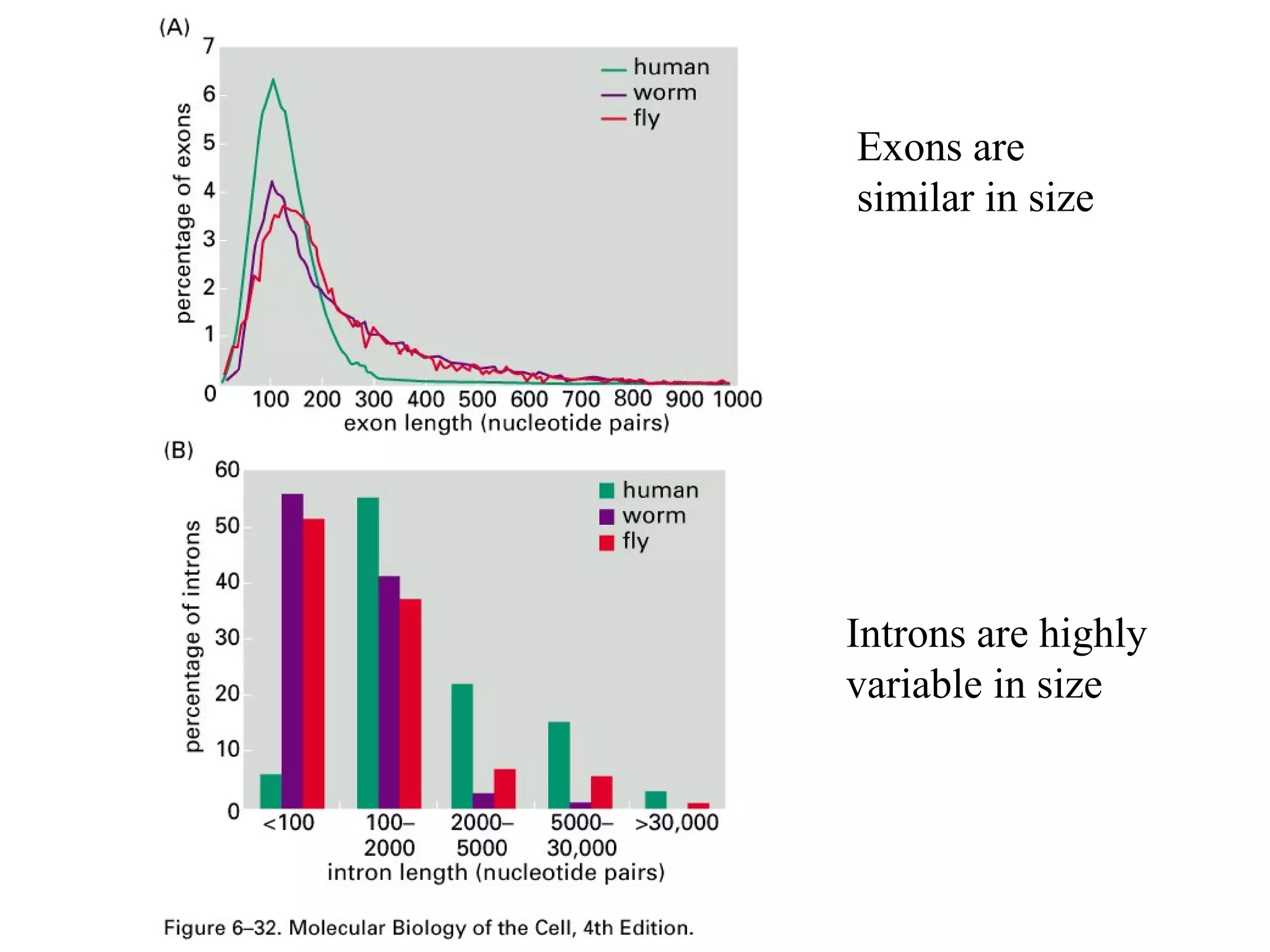
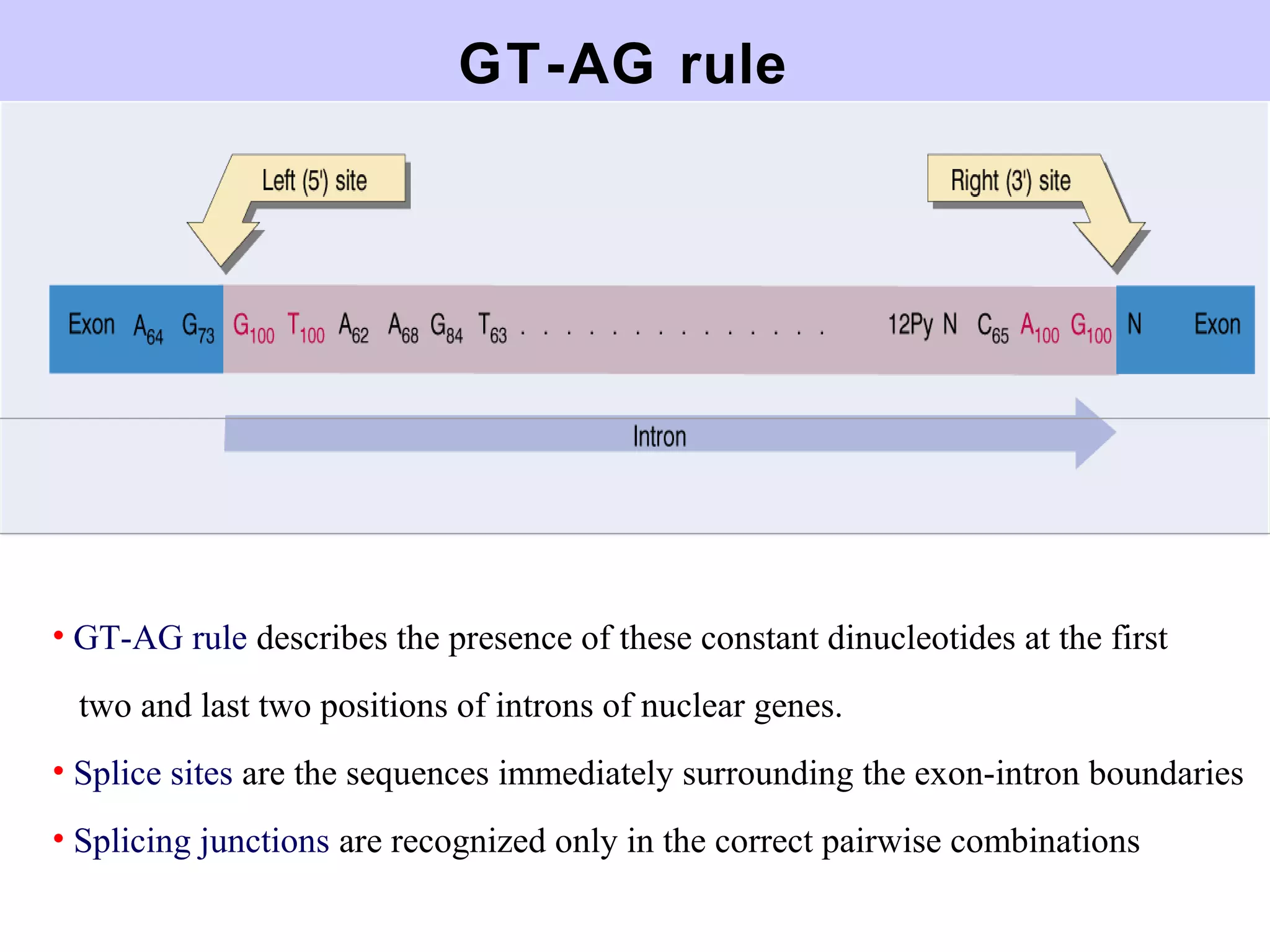
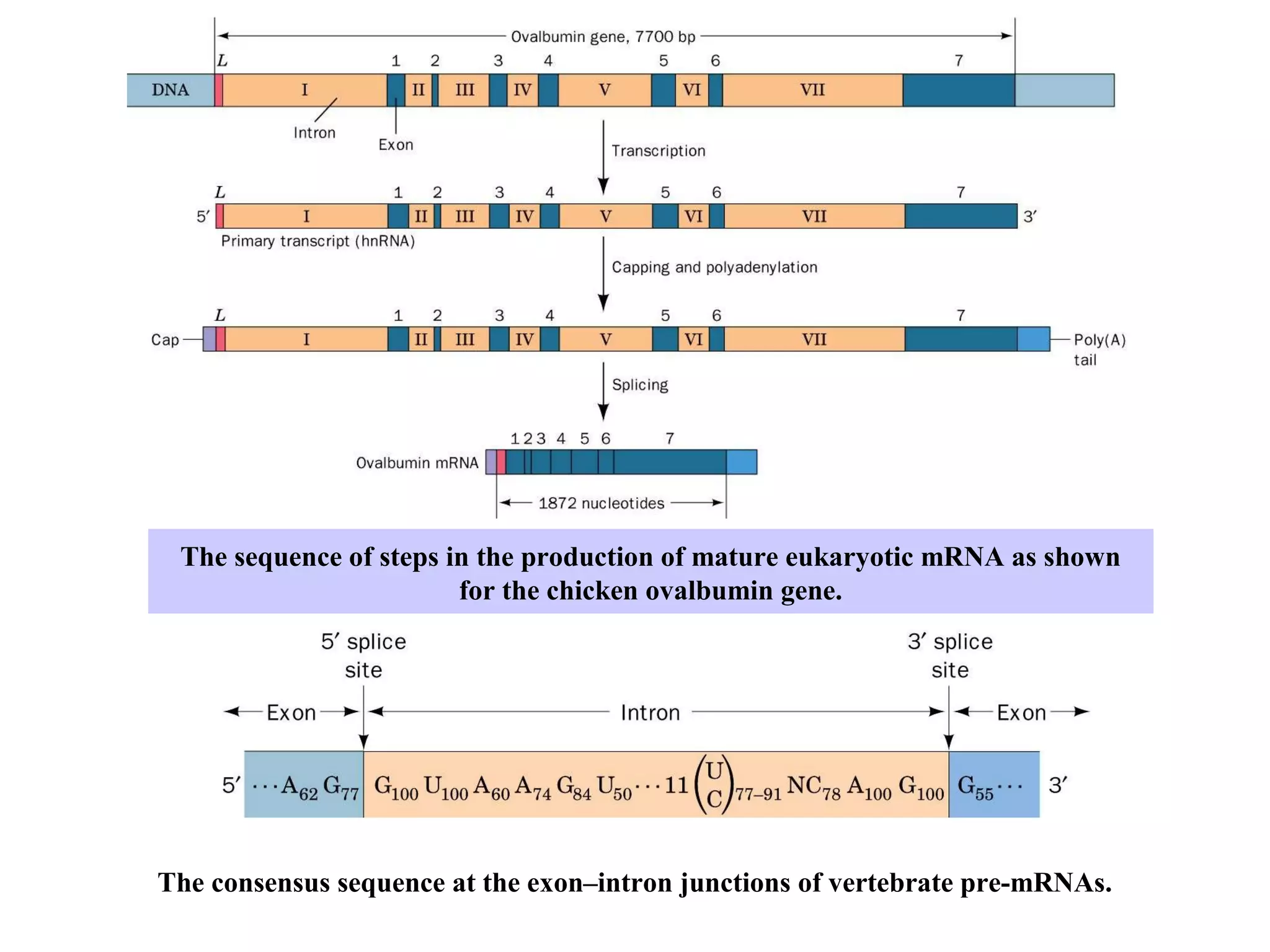
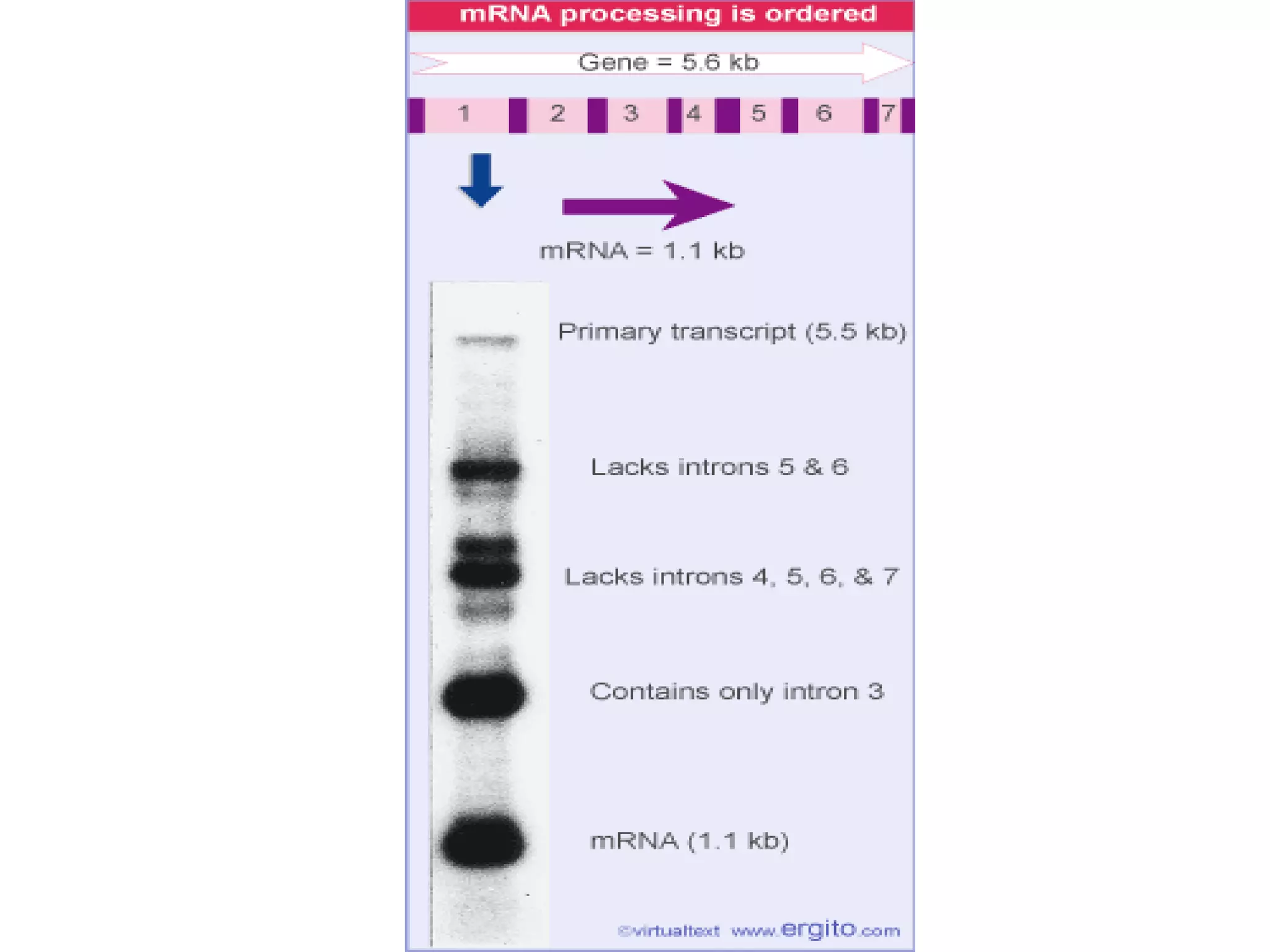
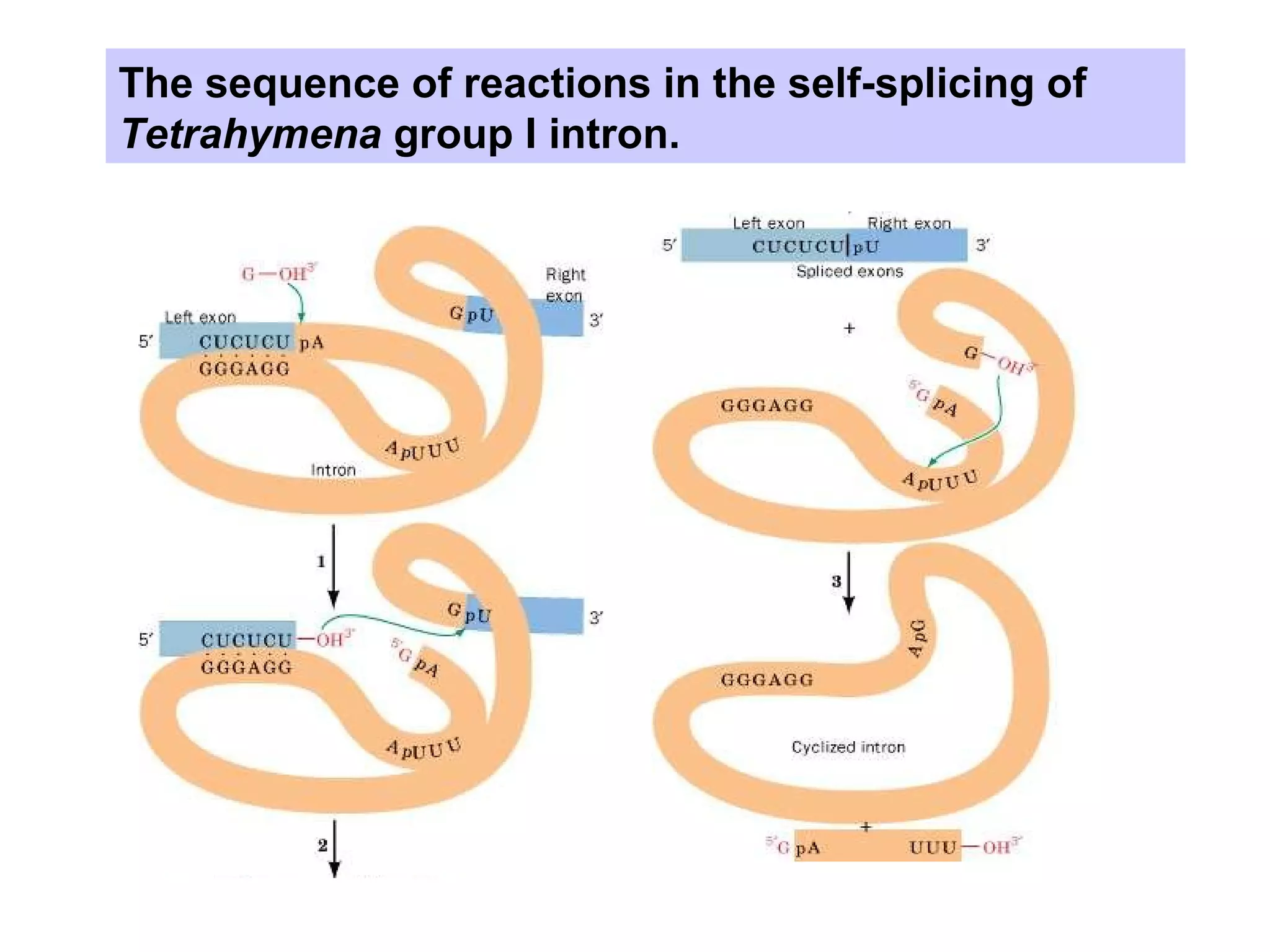
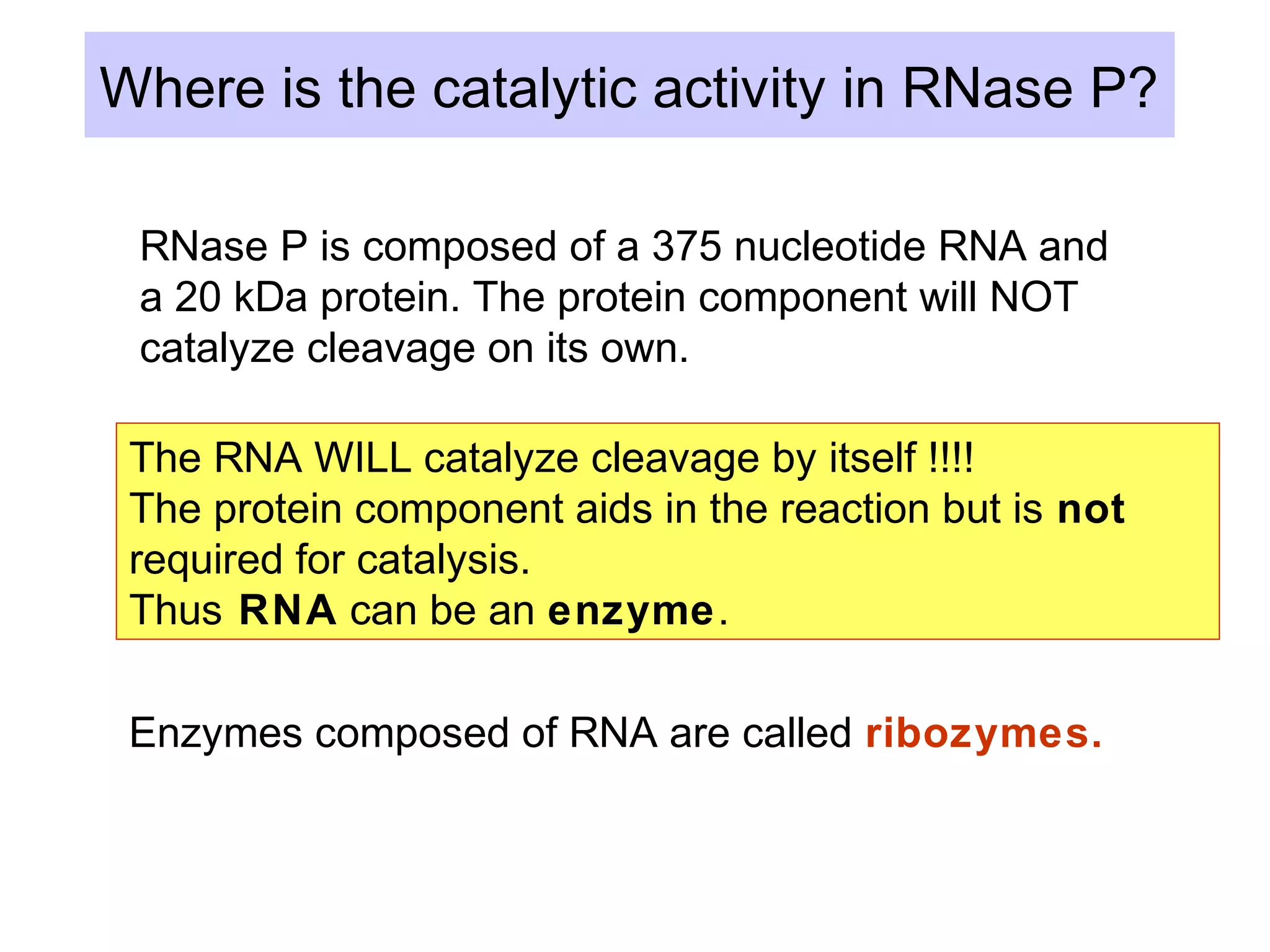
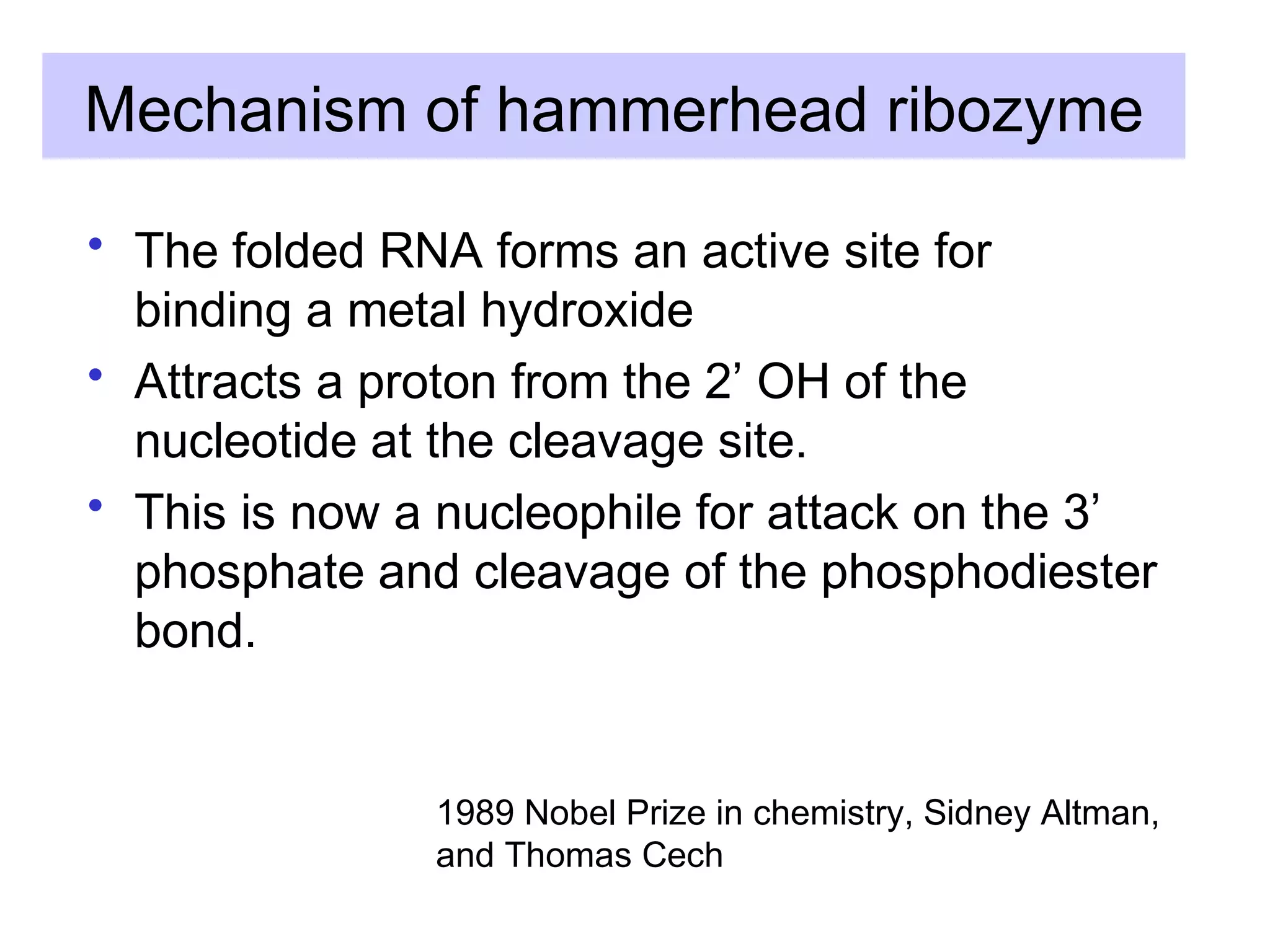
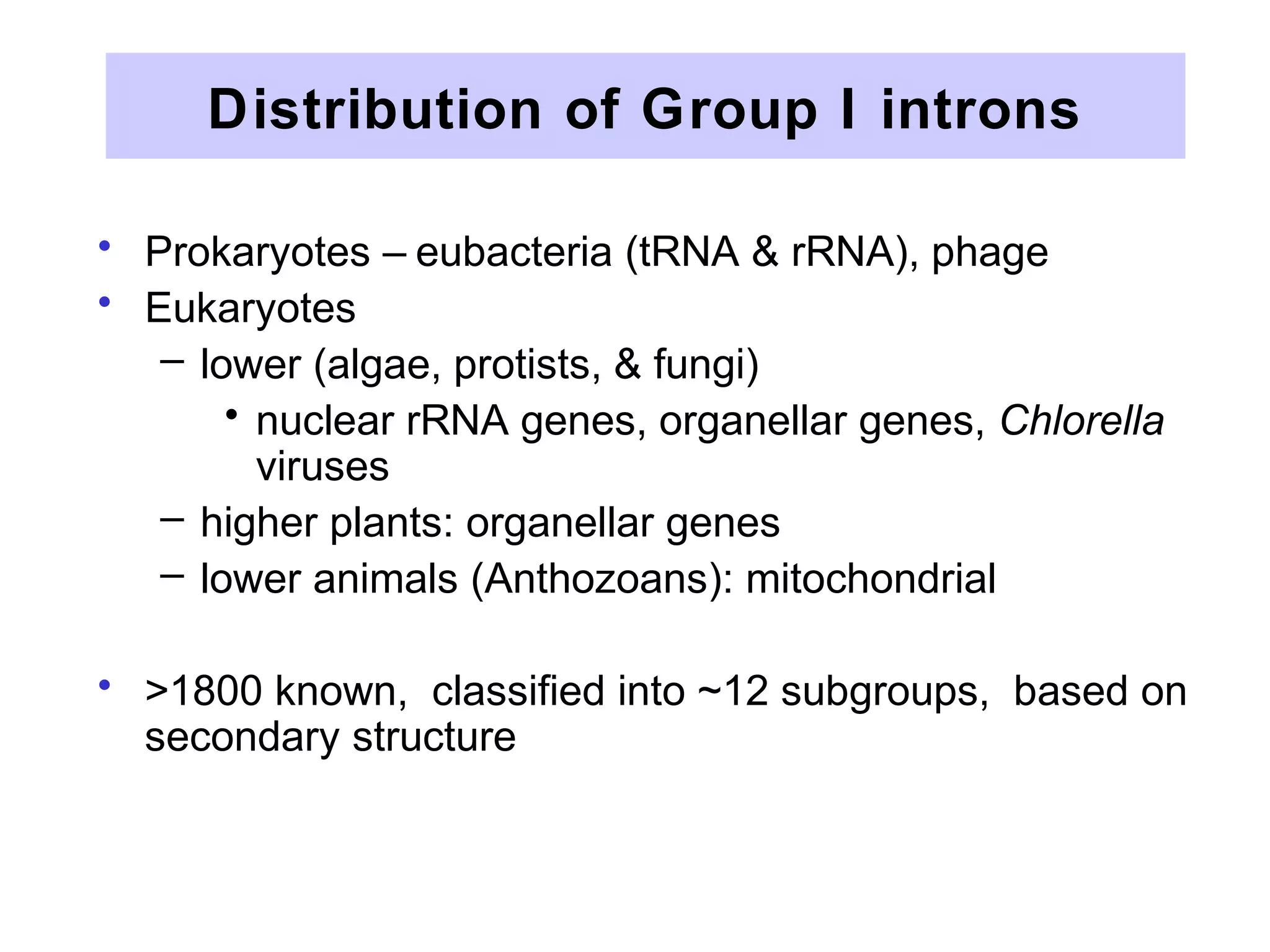
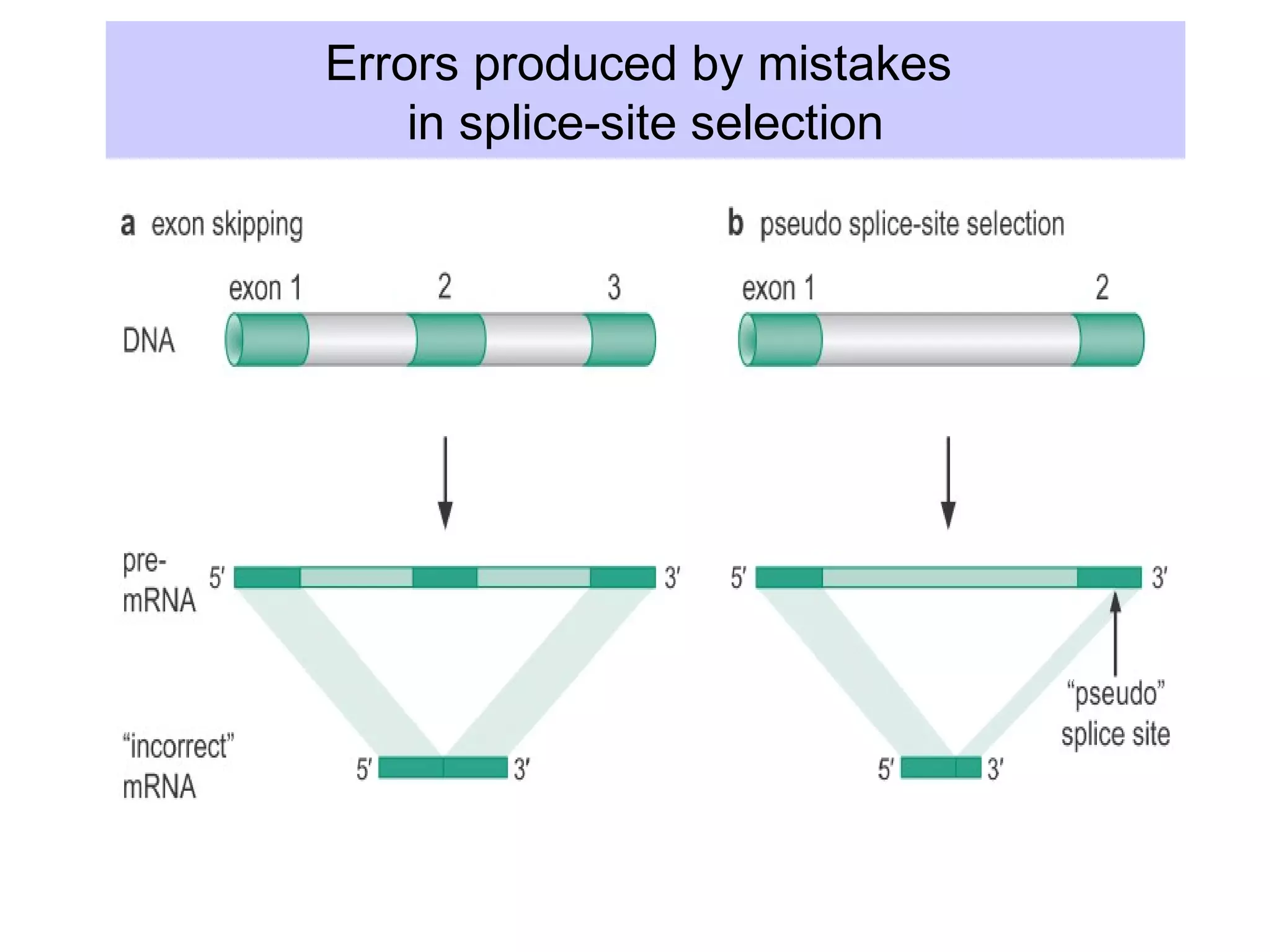
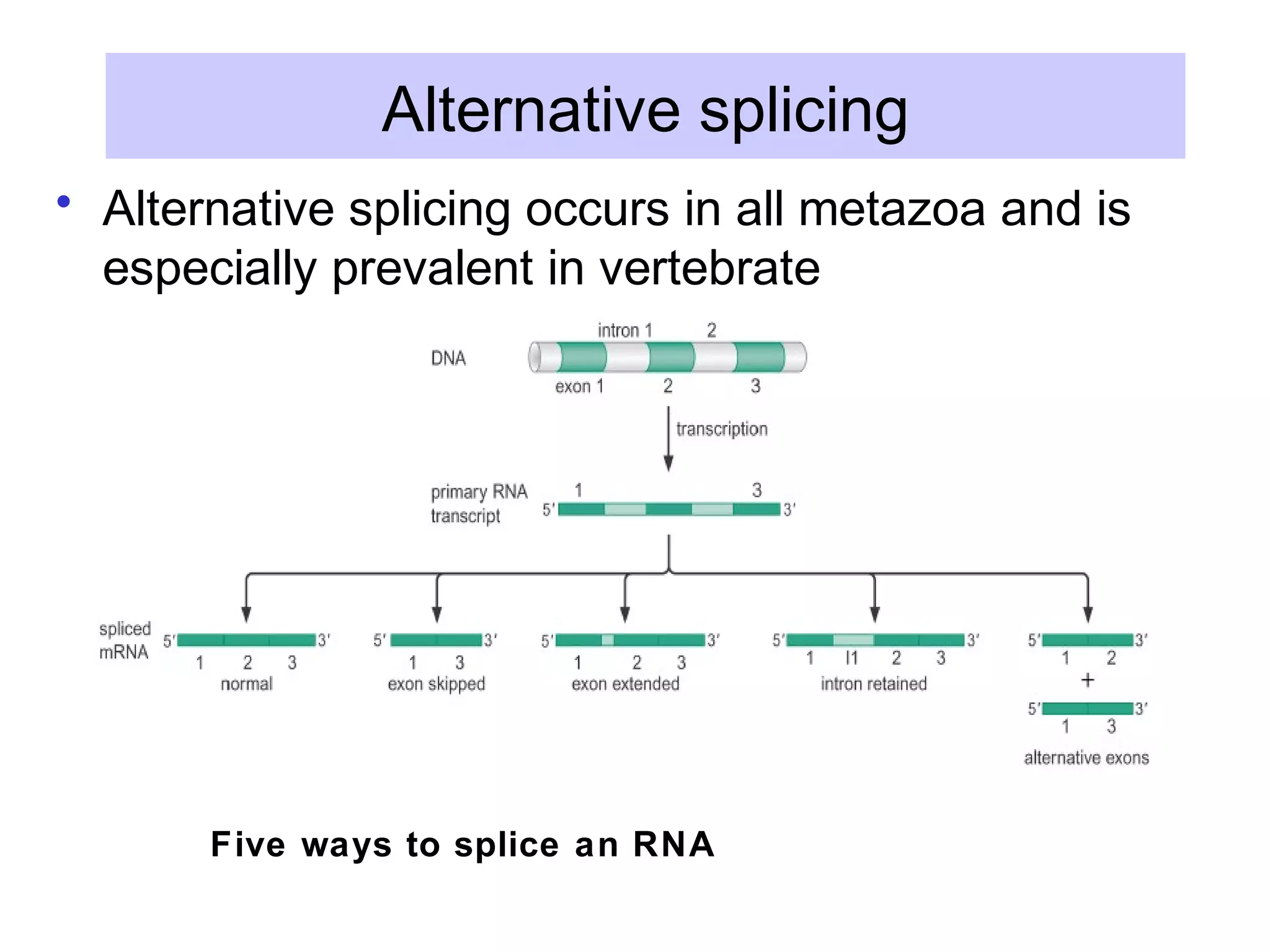
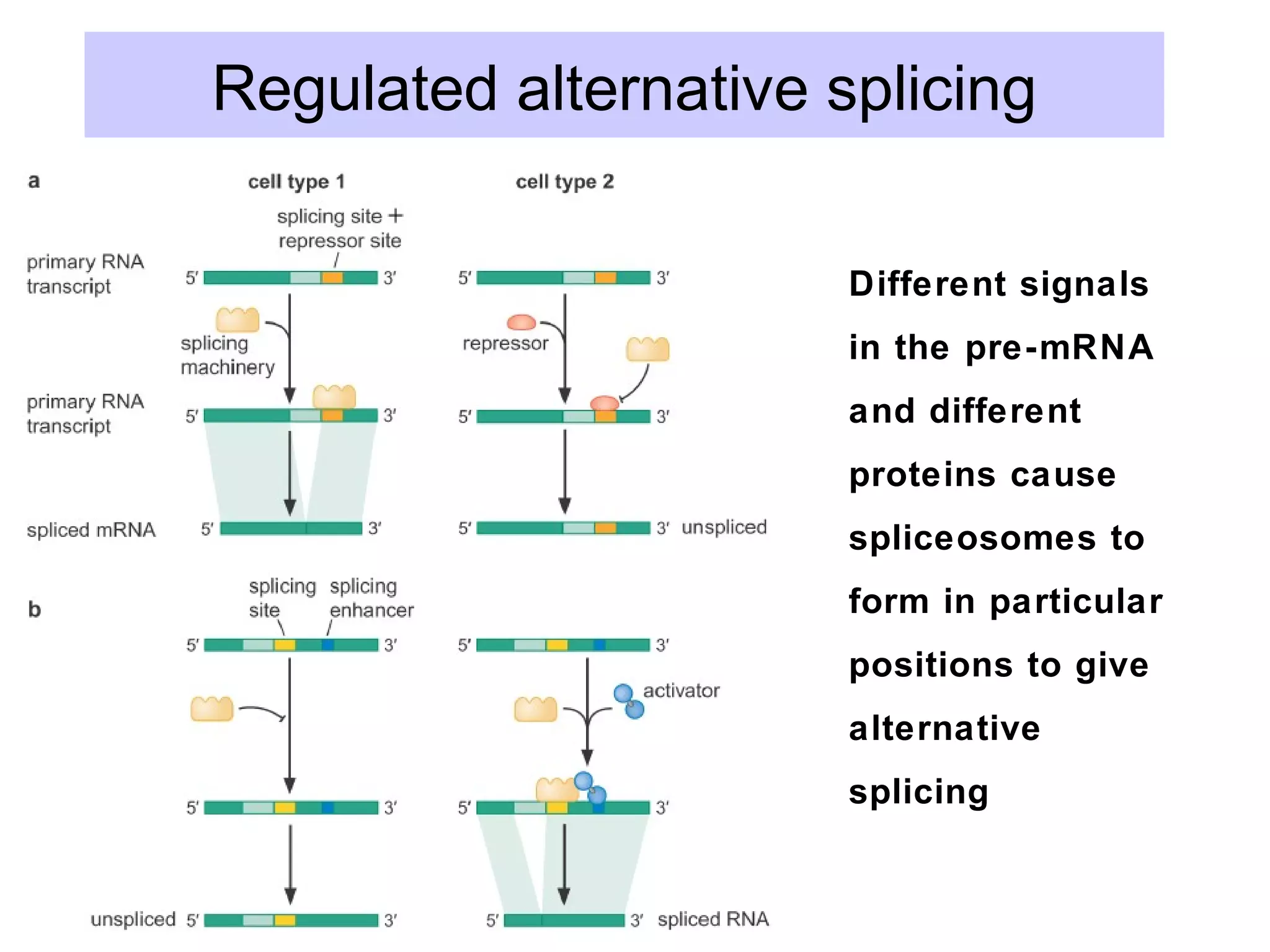
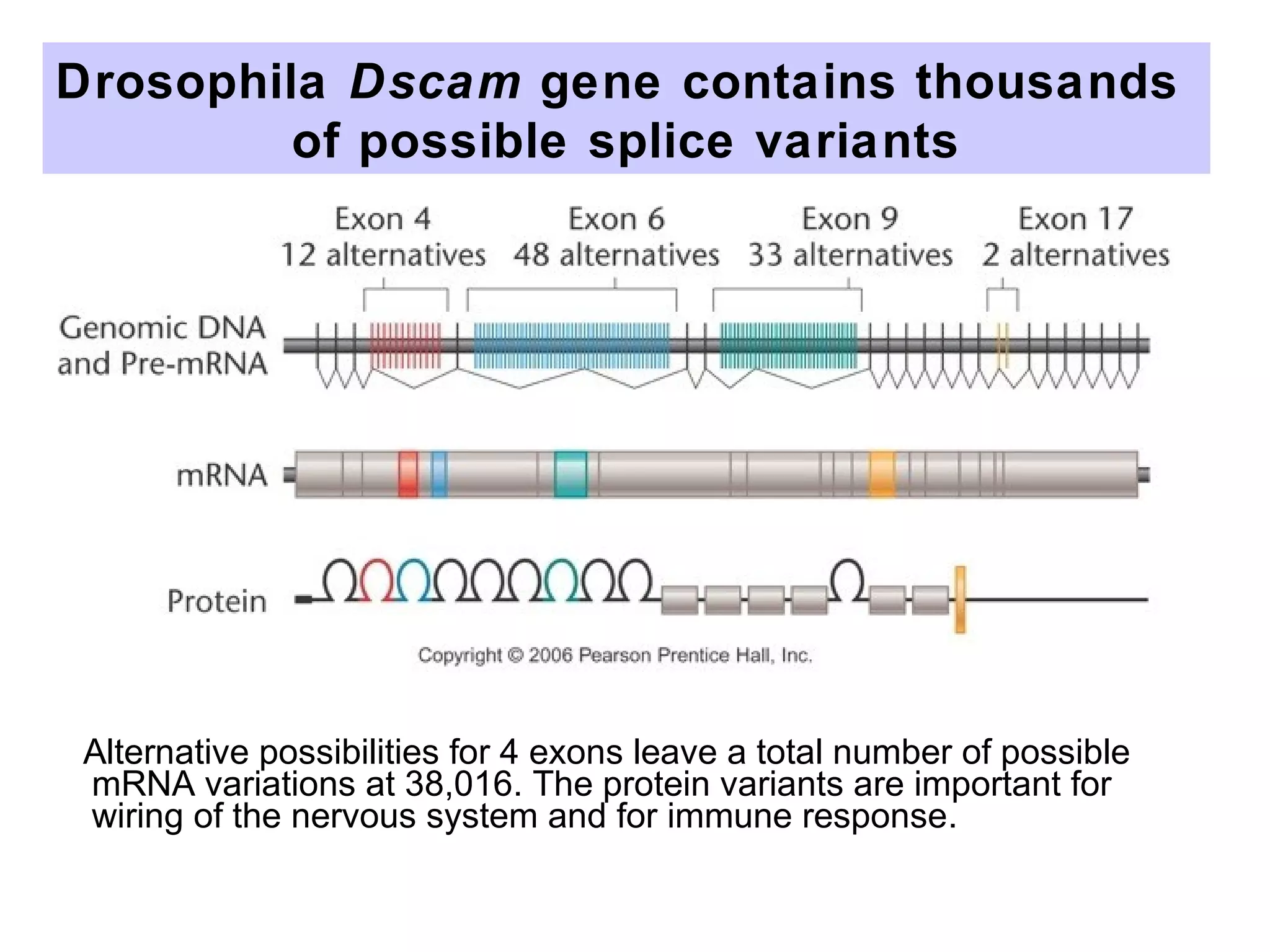
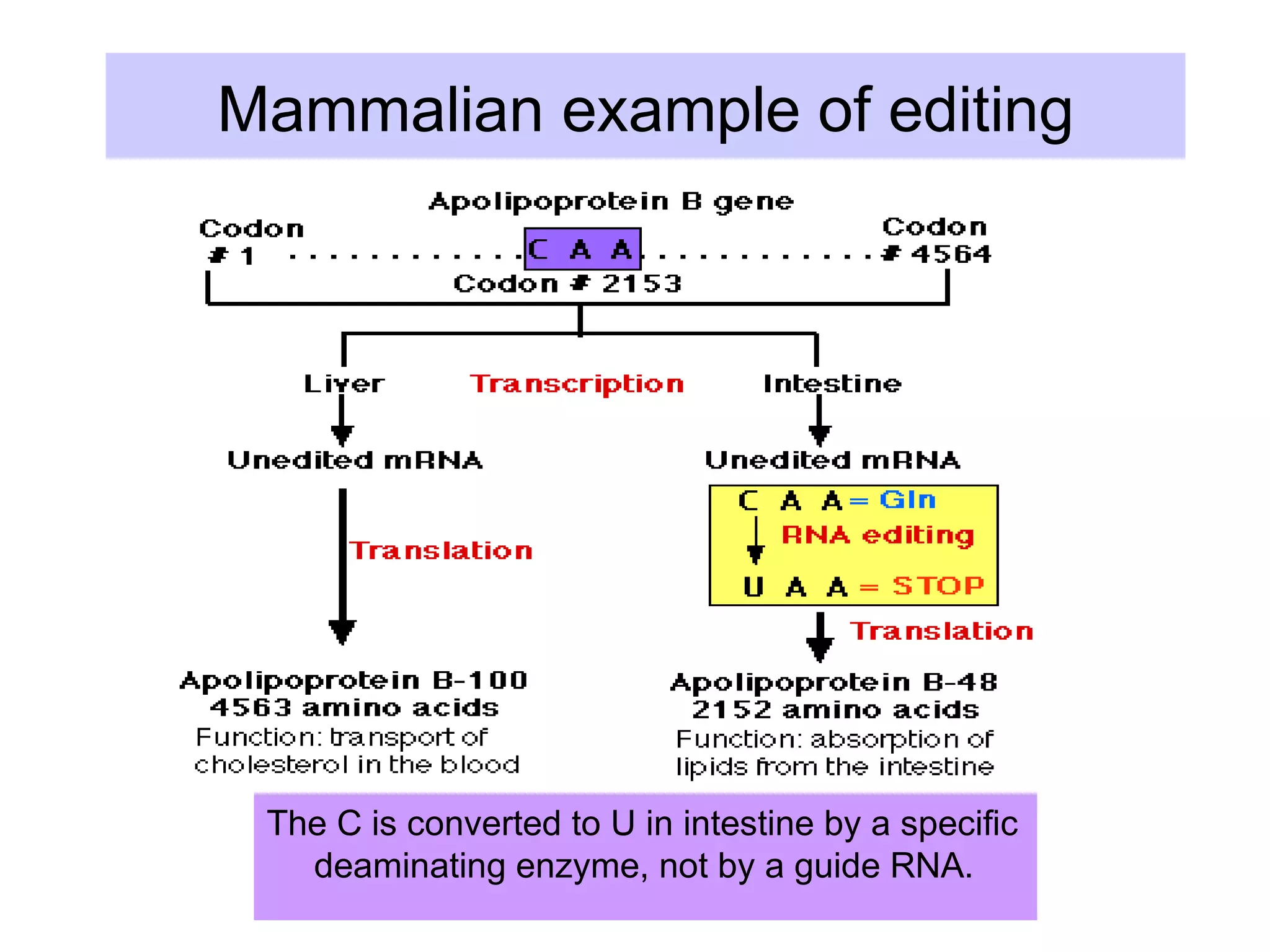
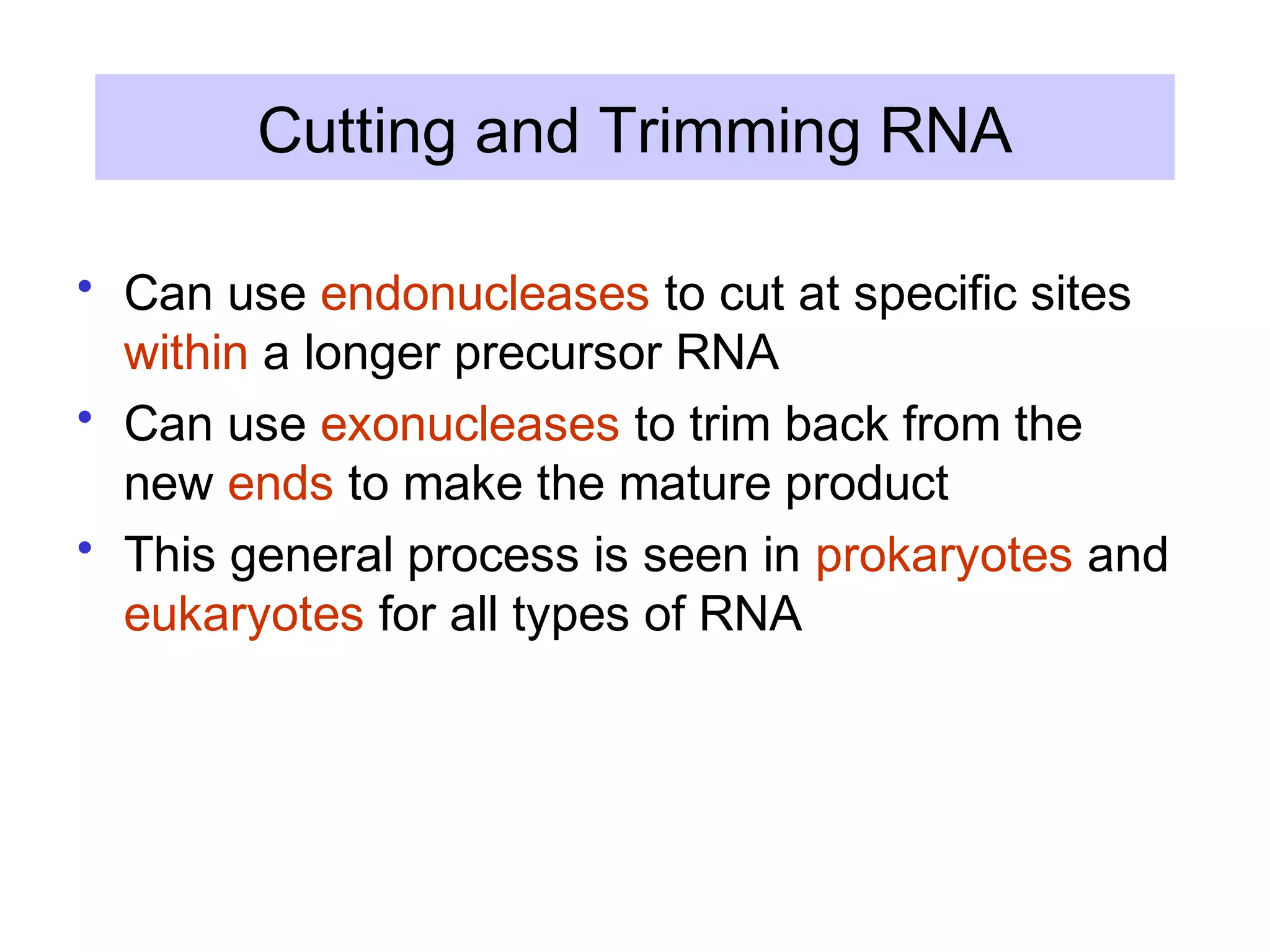
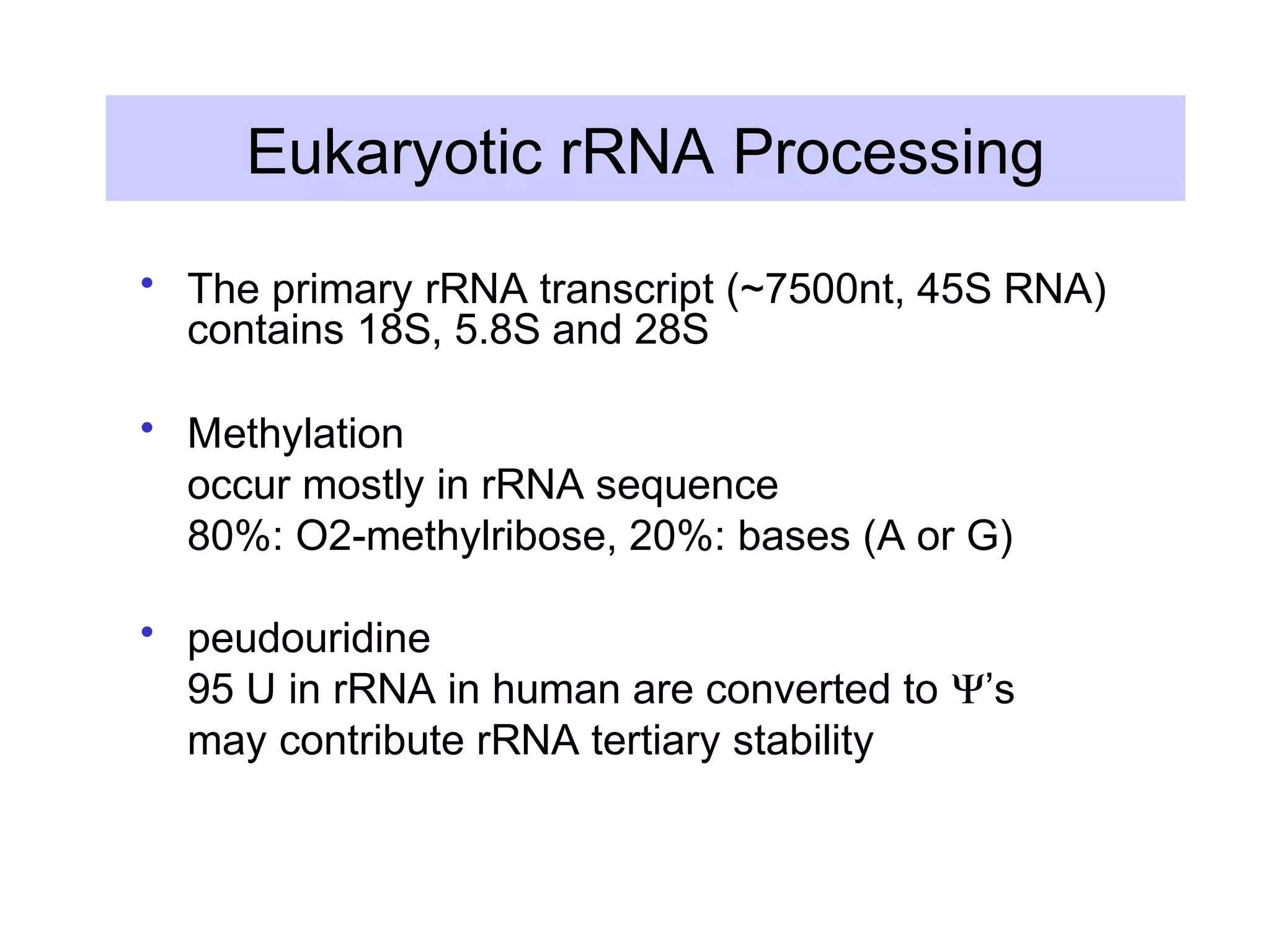
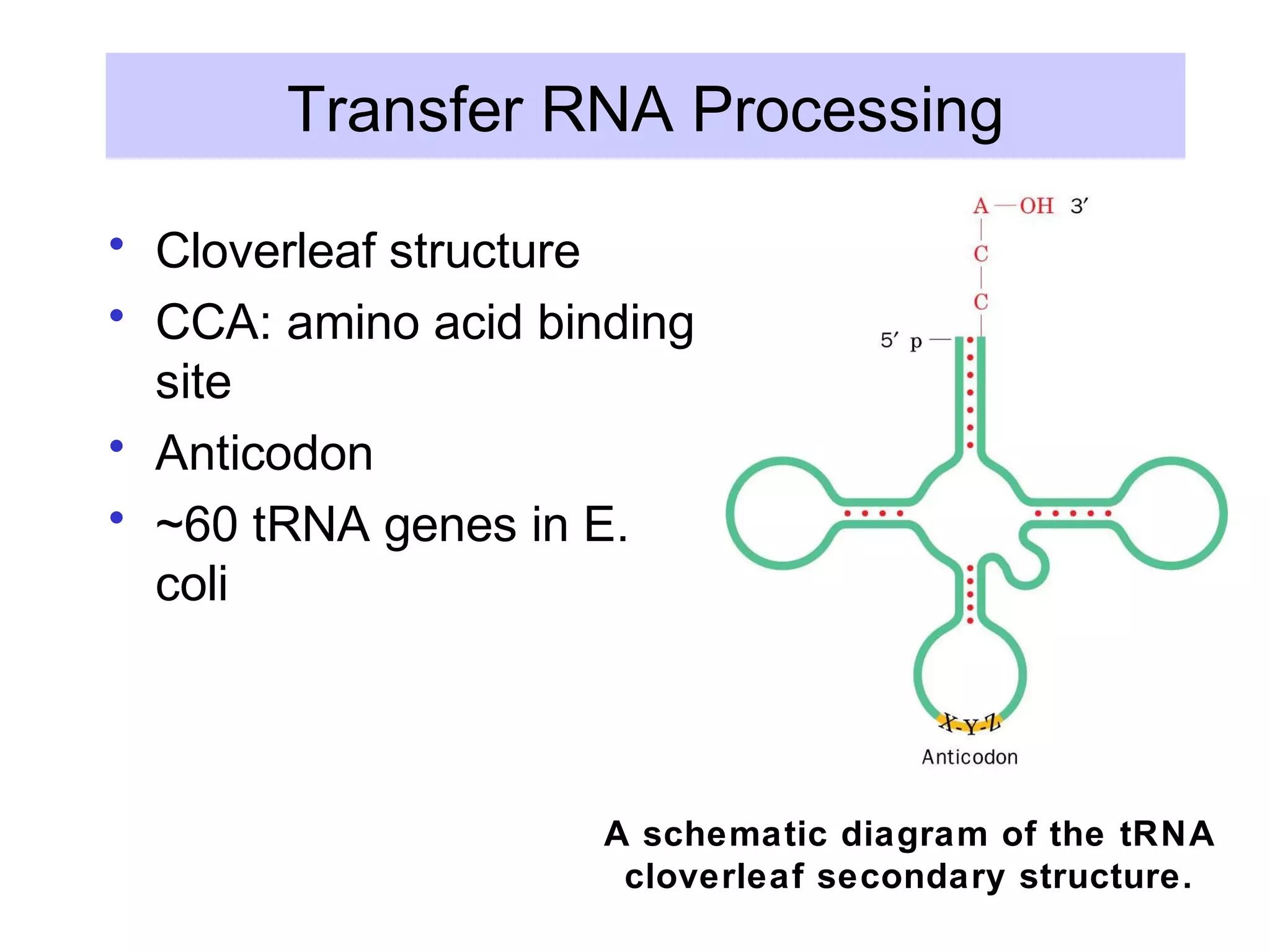
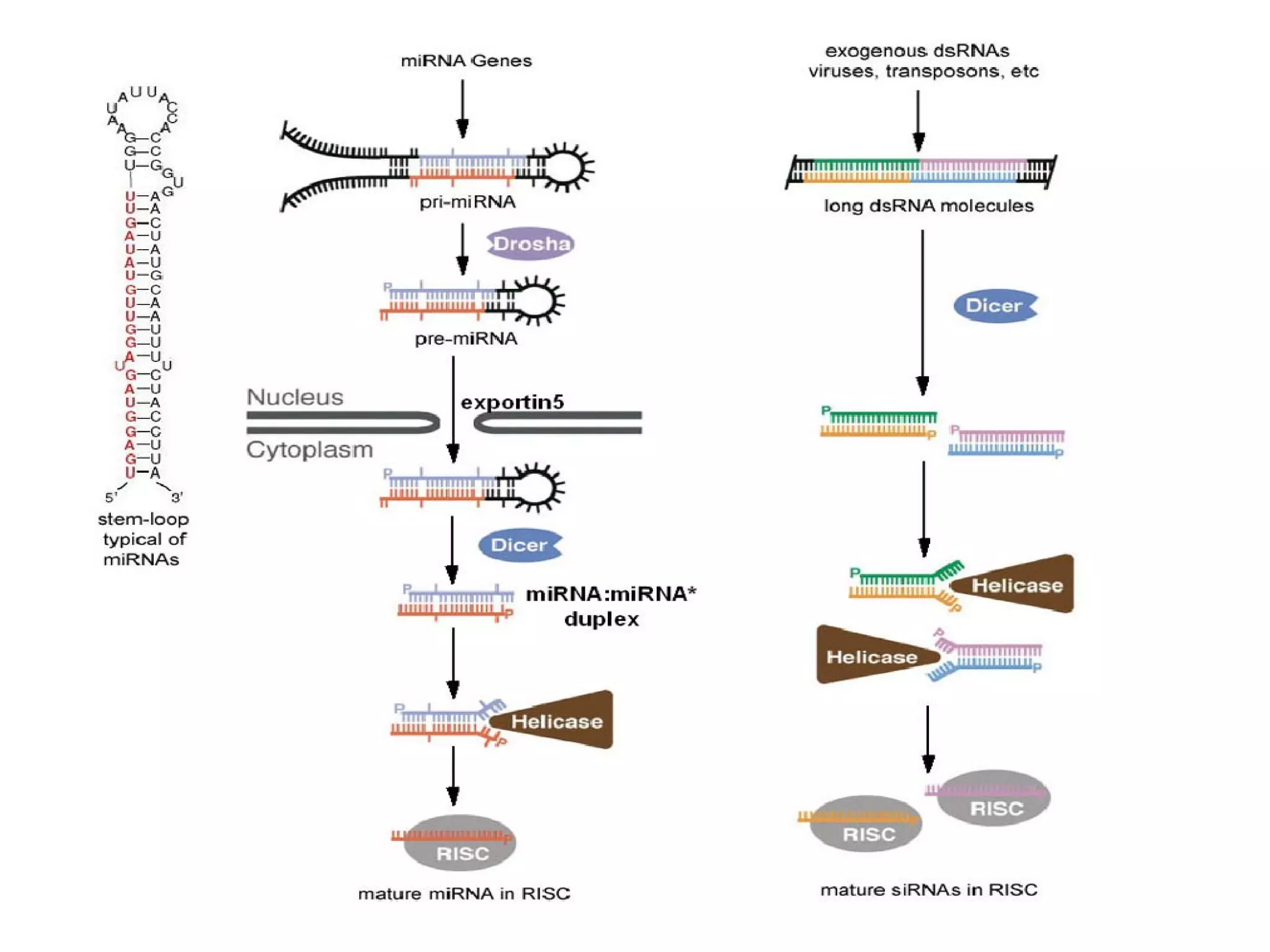
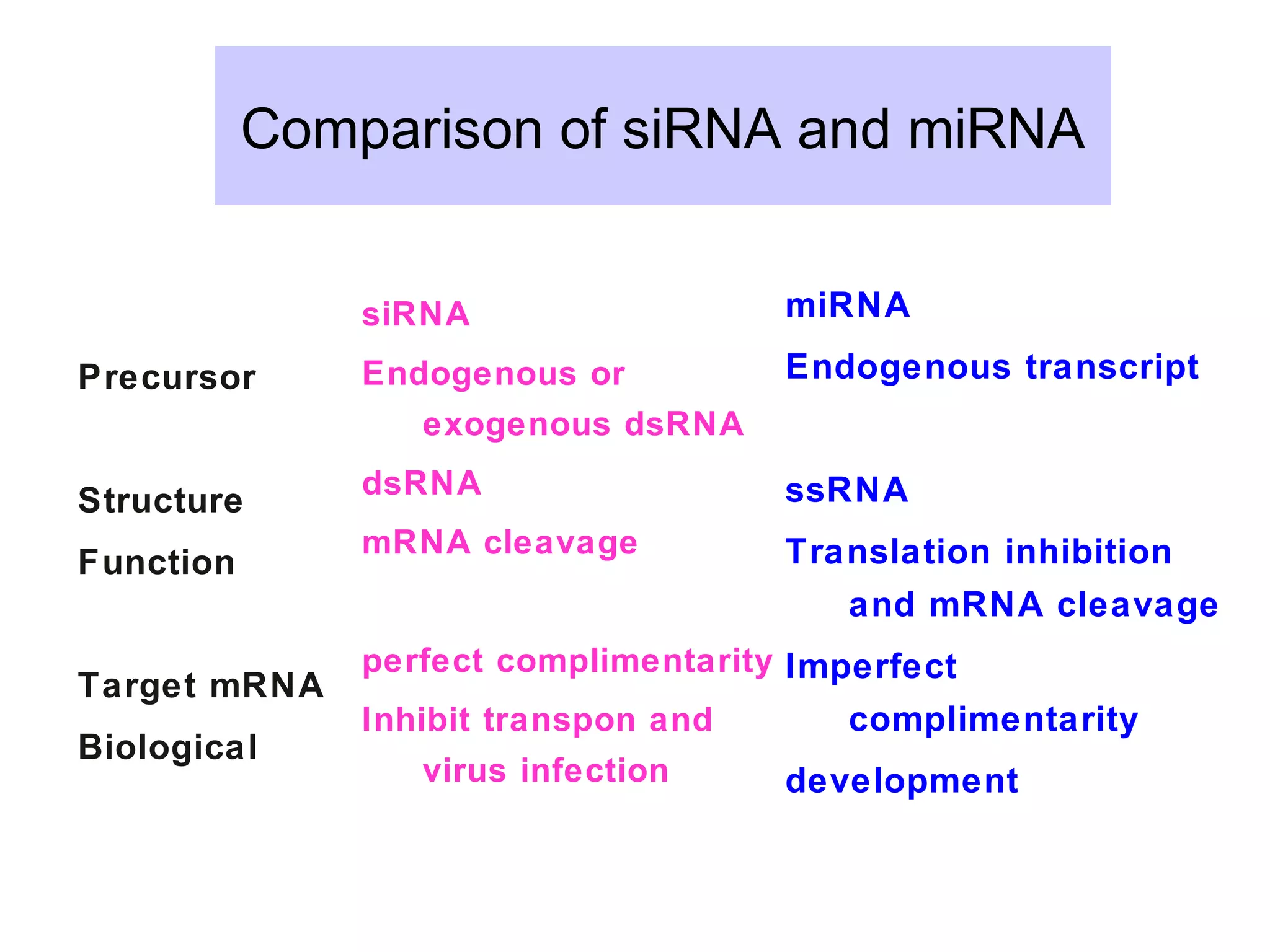
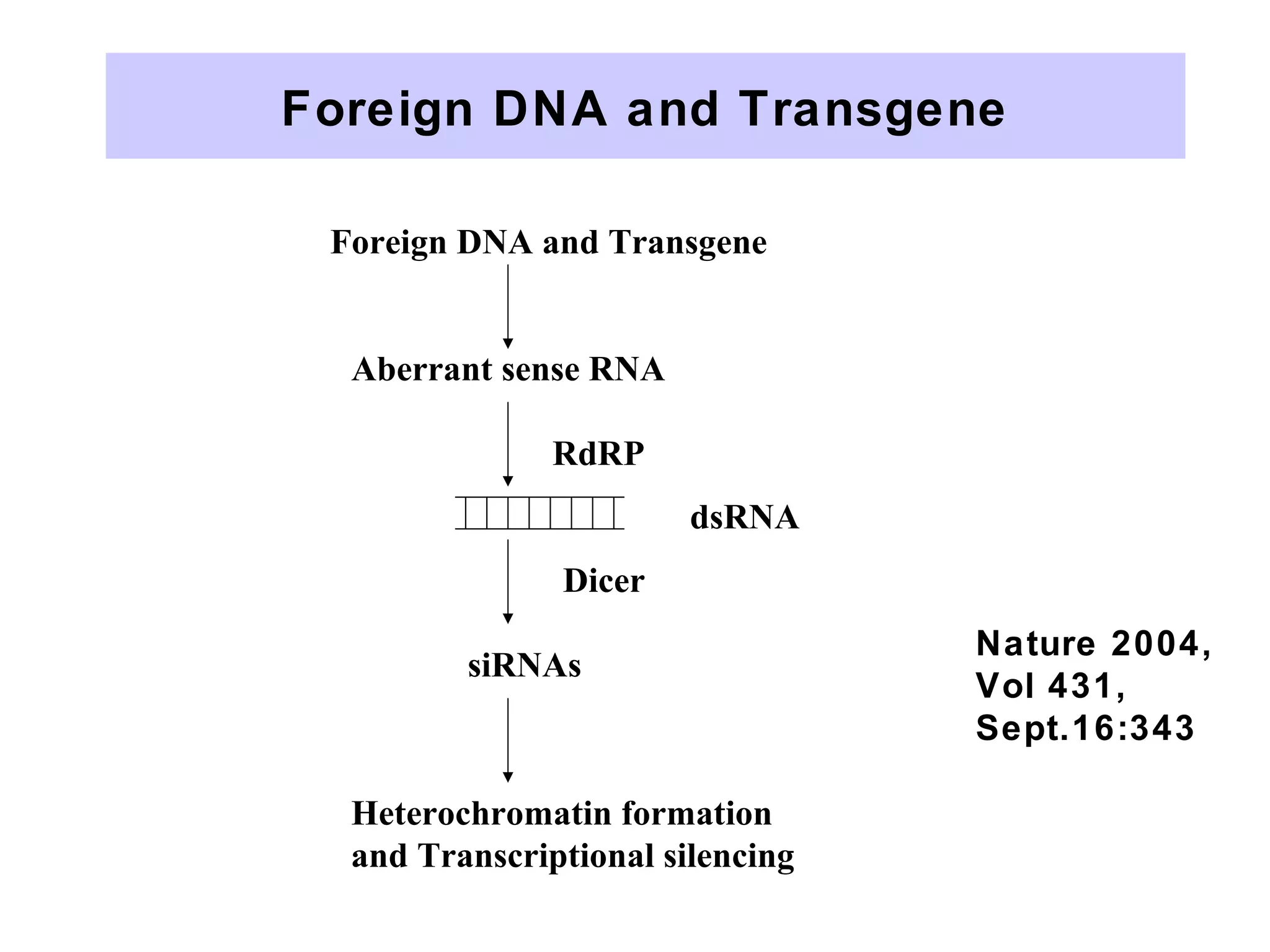
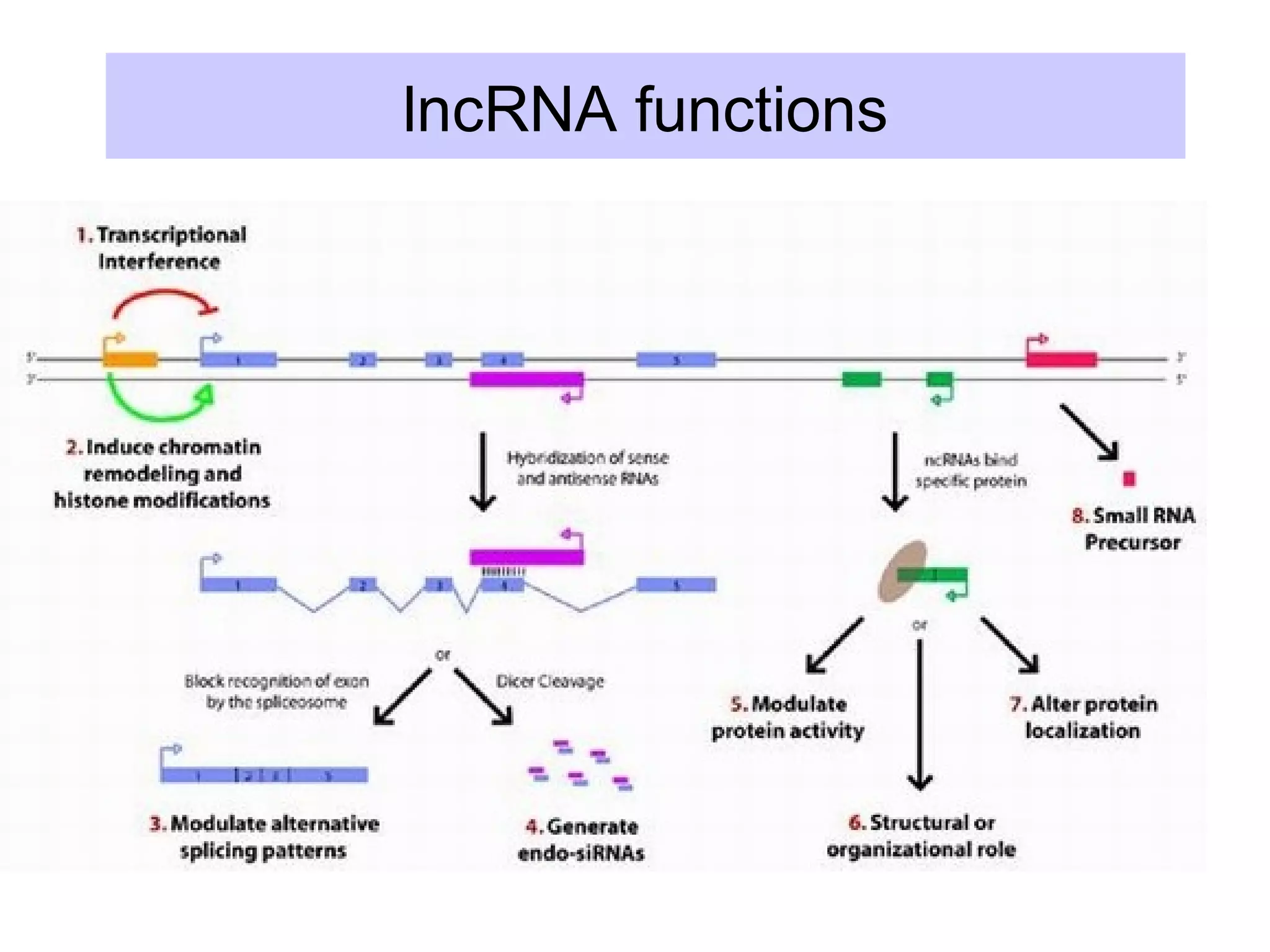
This document summarizes various aspects of post-transcriptional processing of RNA in eukaryotes. It discusses:
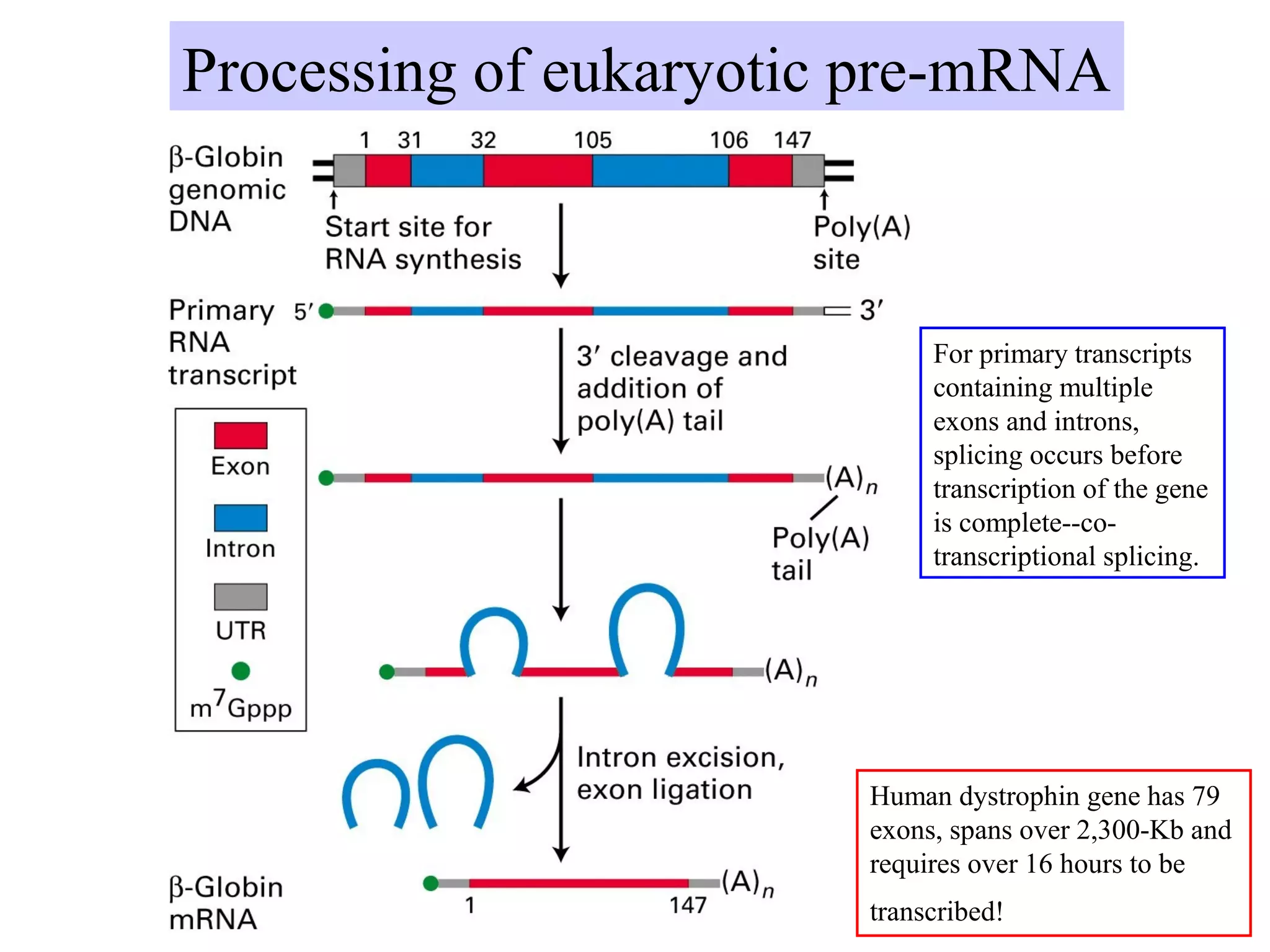
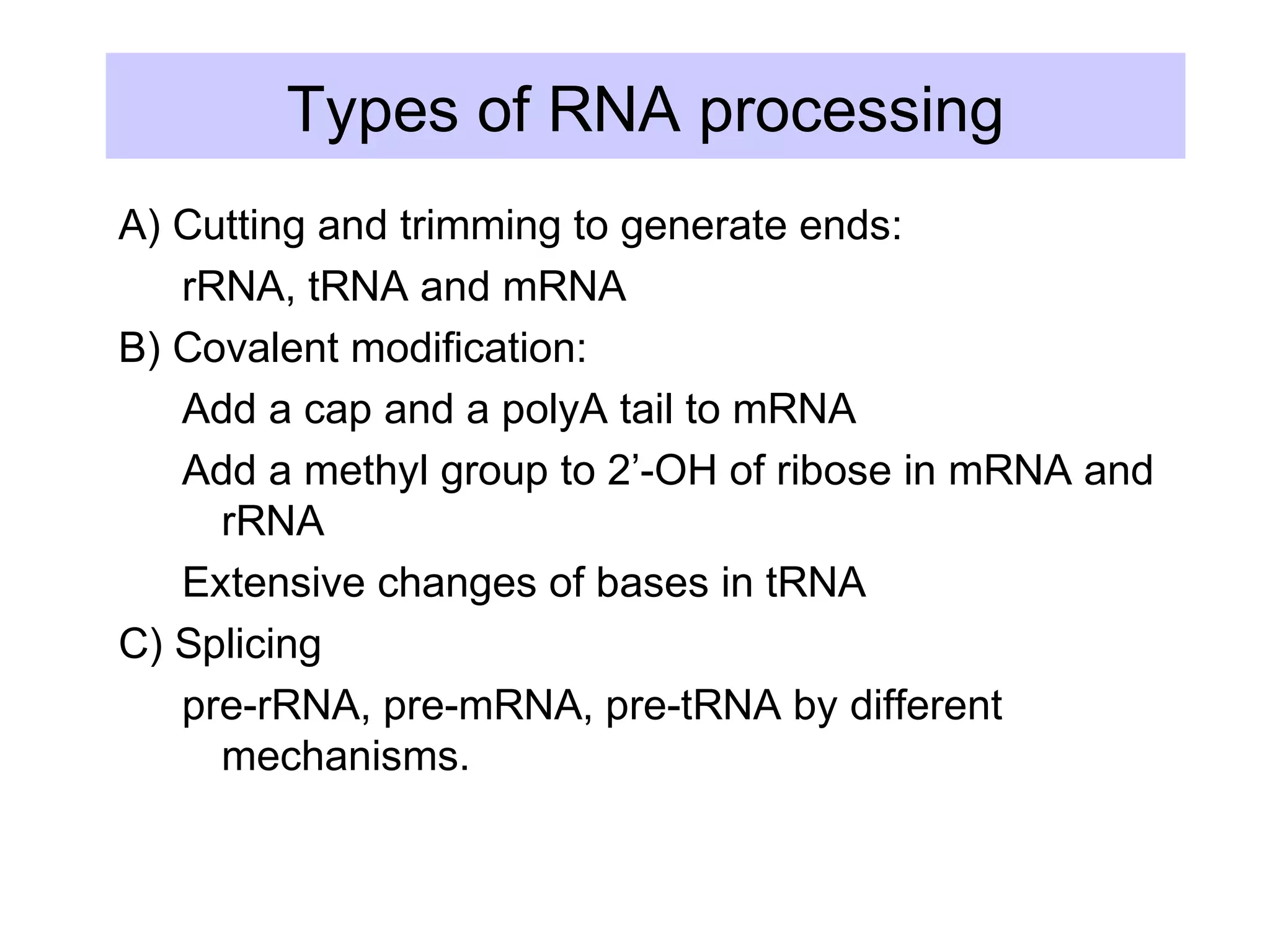
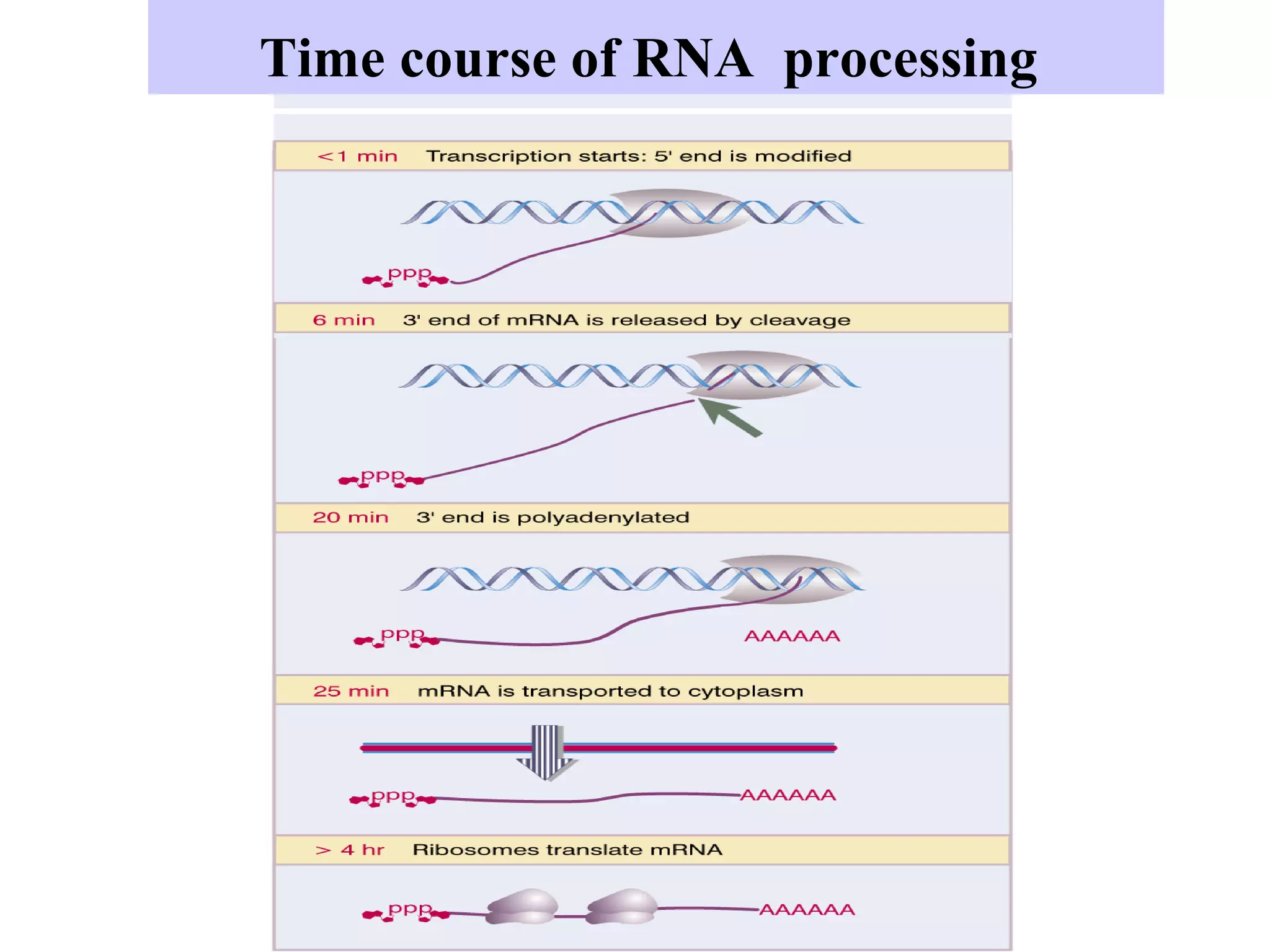
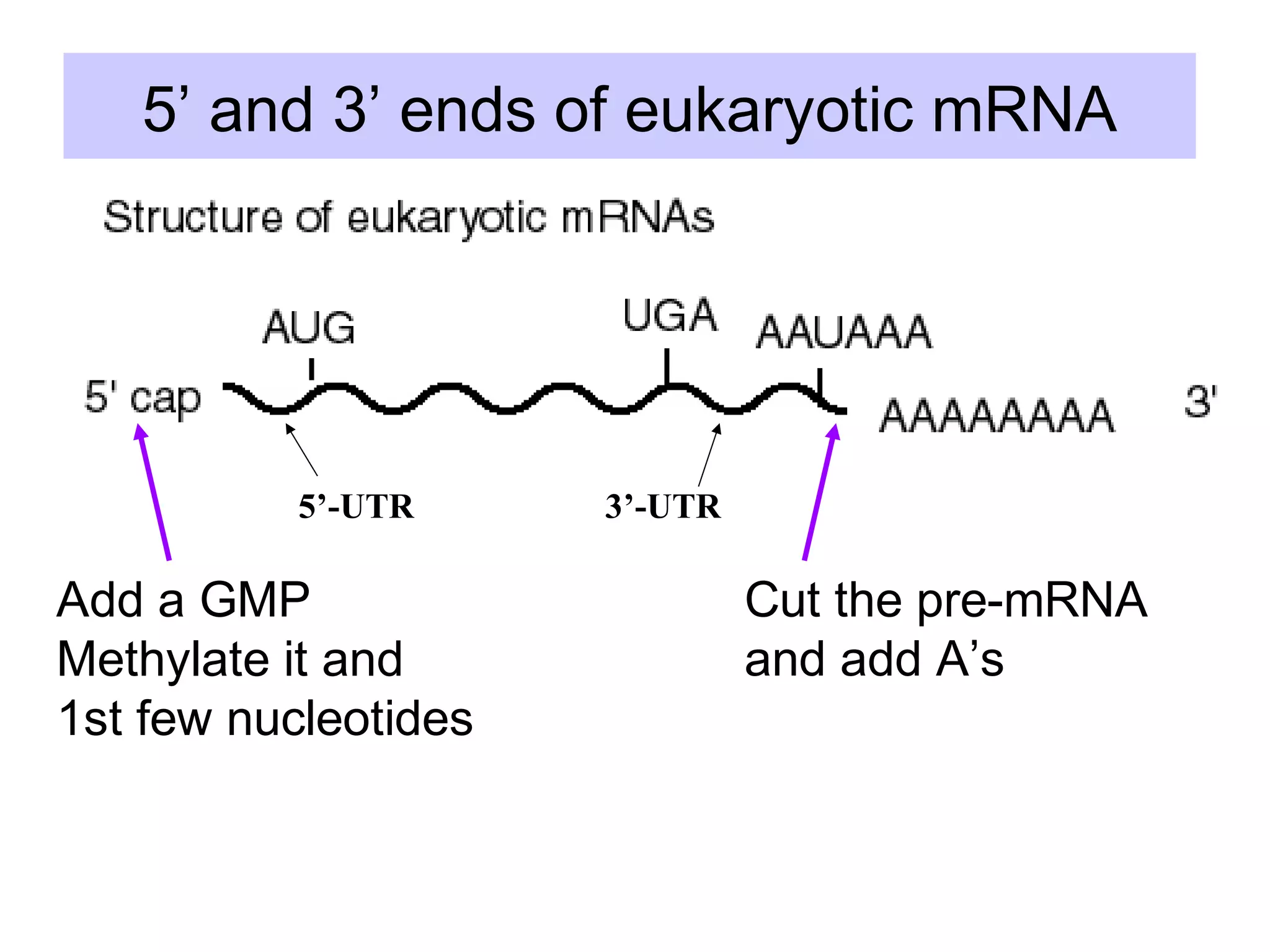
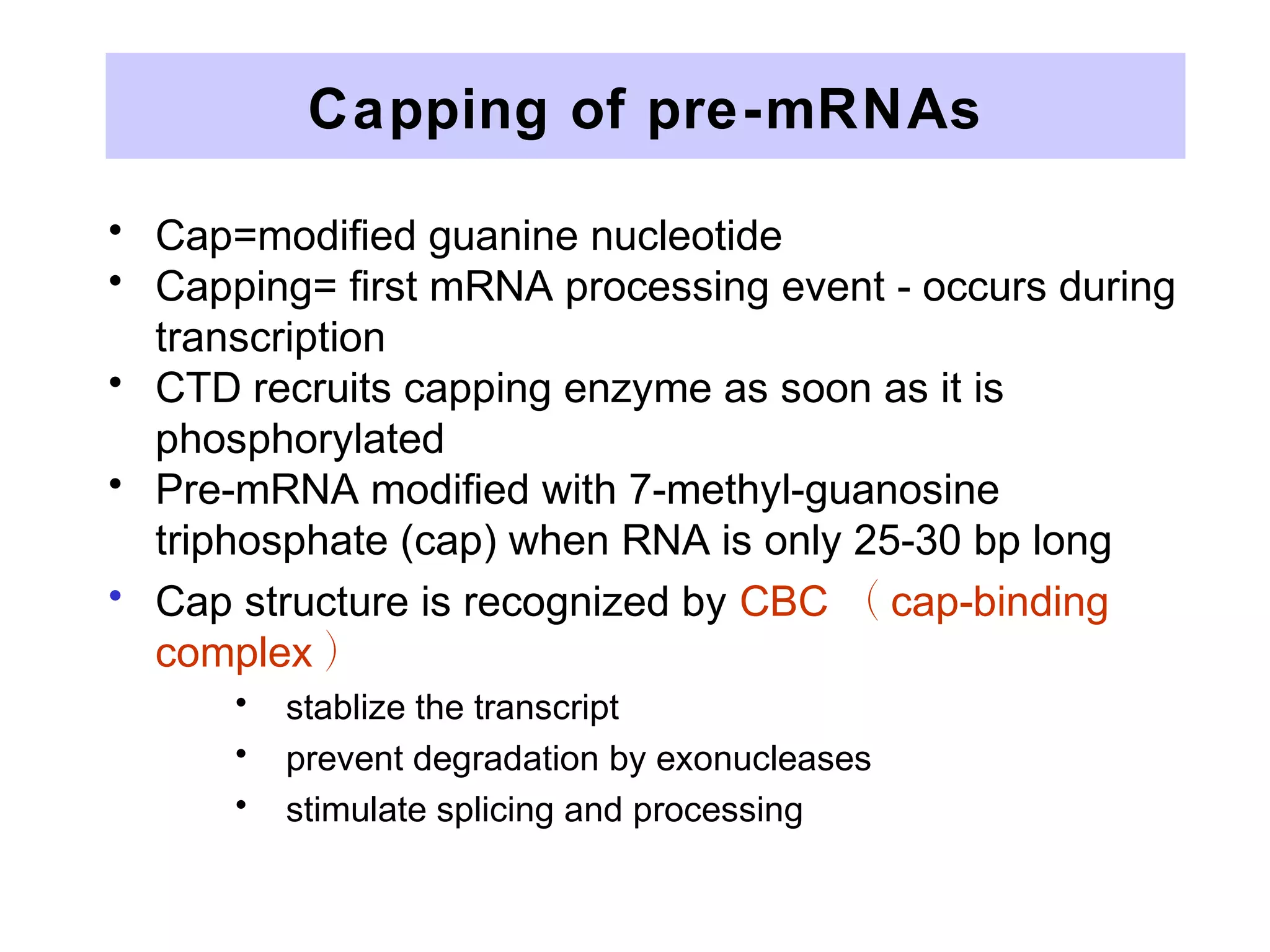
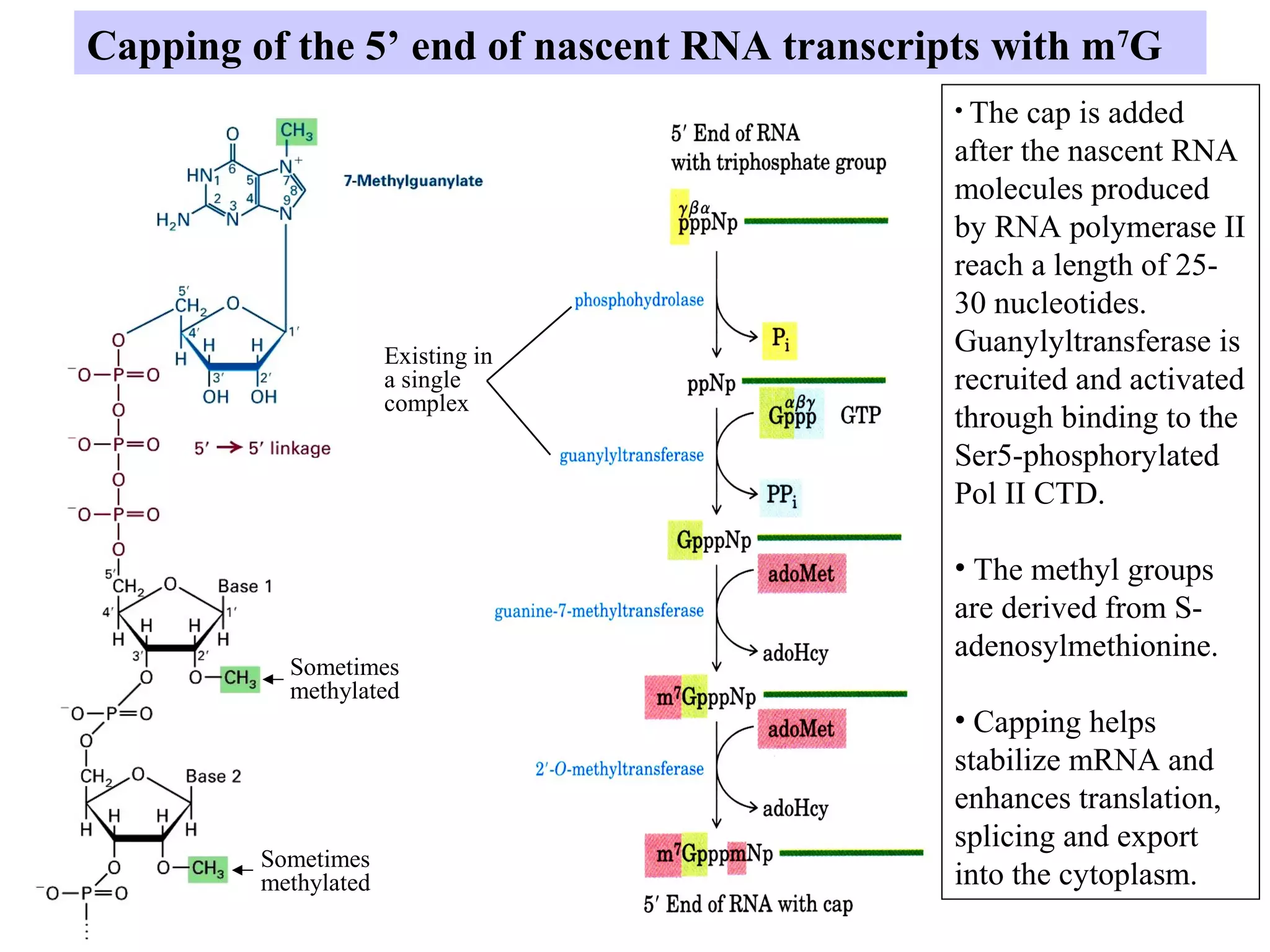
1) Primary transcripts (hnRNA) that are processed in the nucleus before being exported to the cytoplasm. Processing includes 5' capping, splicing of introns, and 3' polyadenylation.
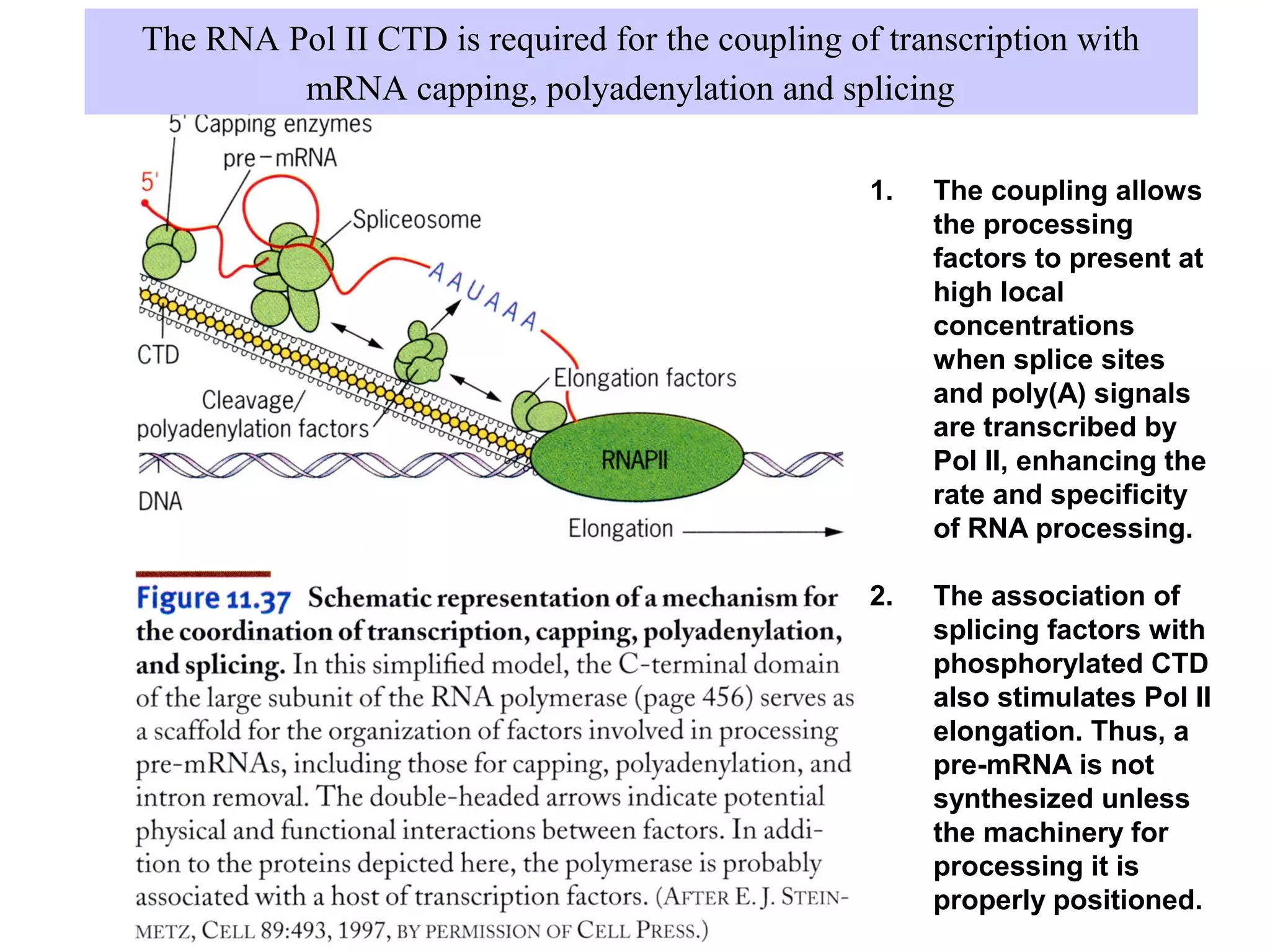
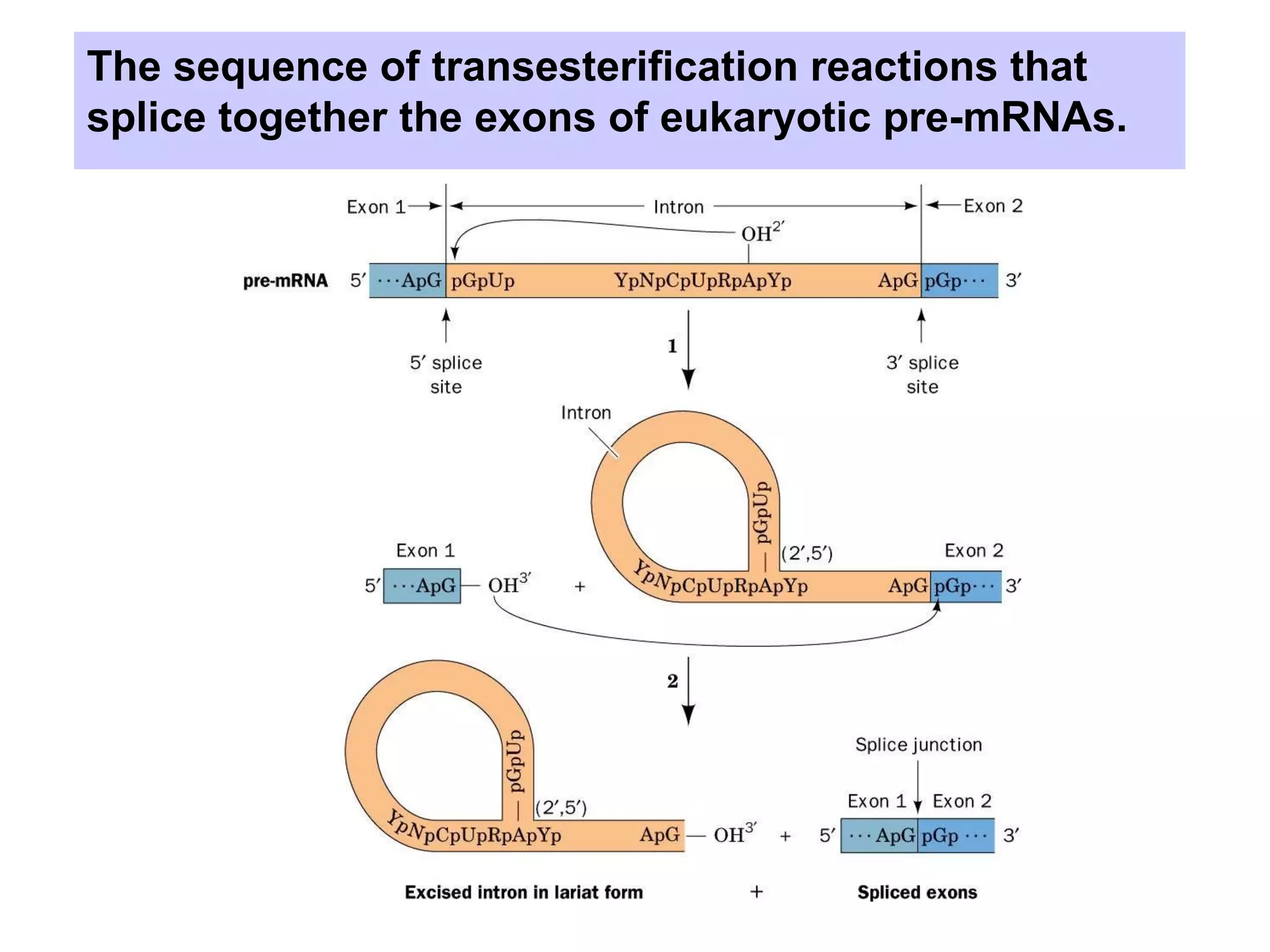
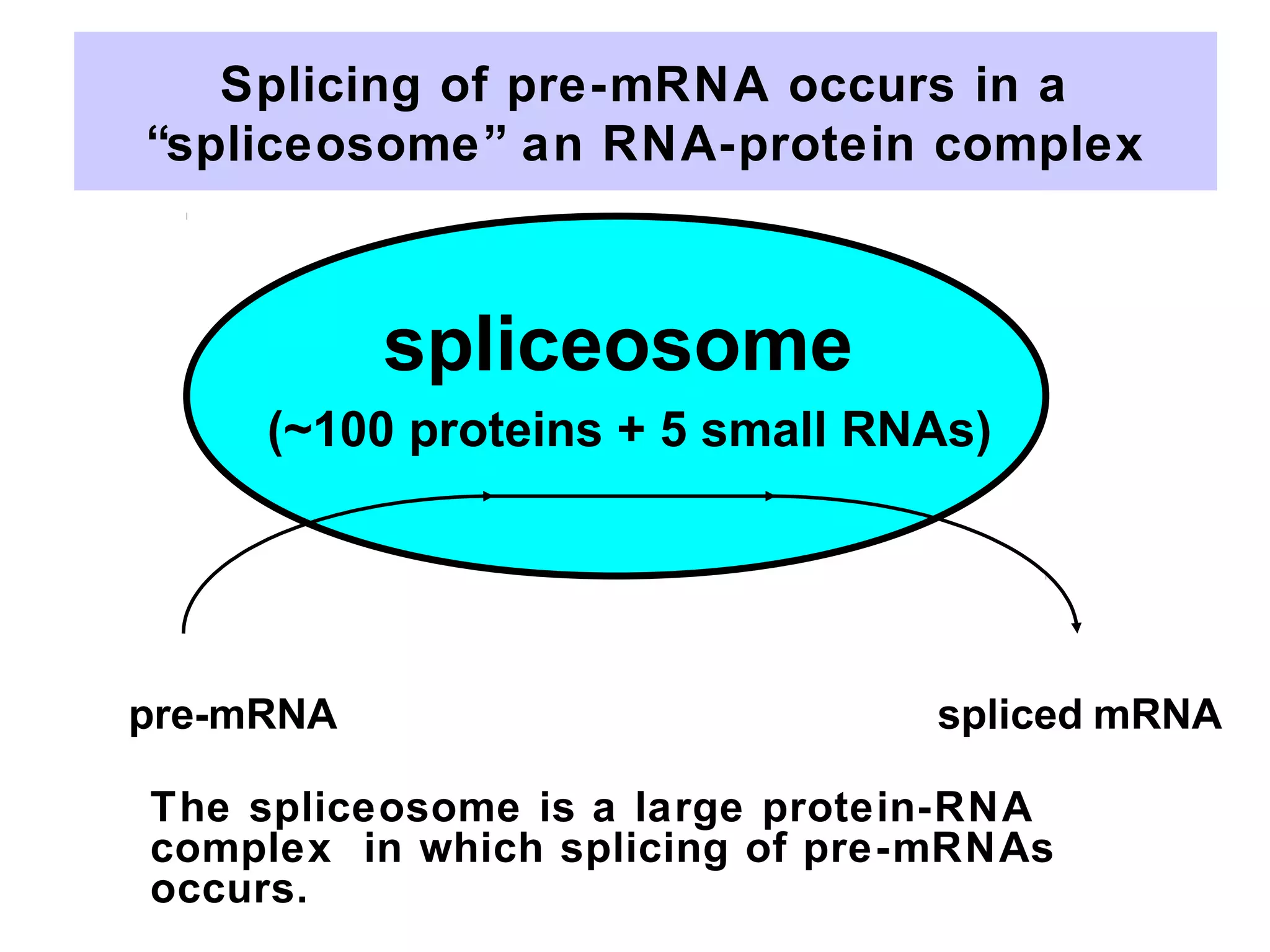
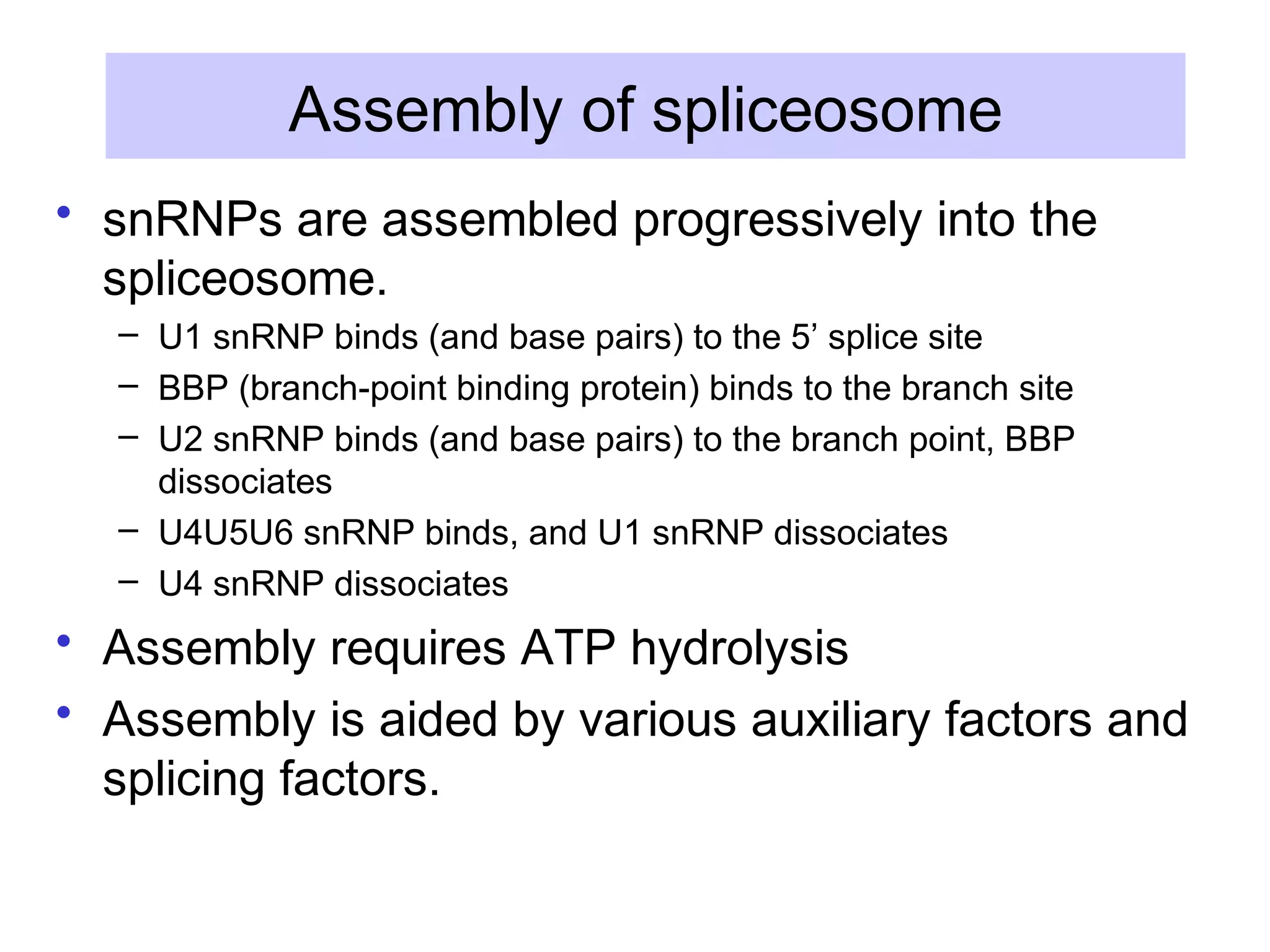
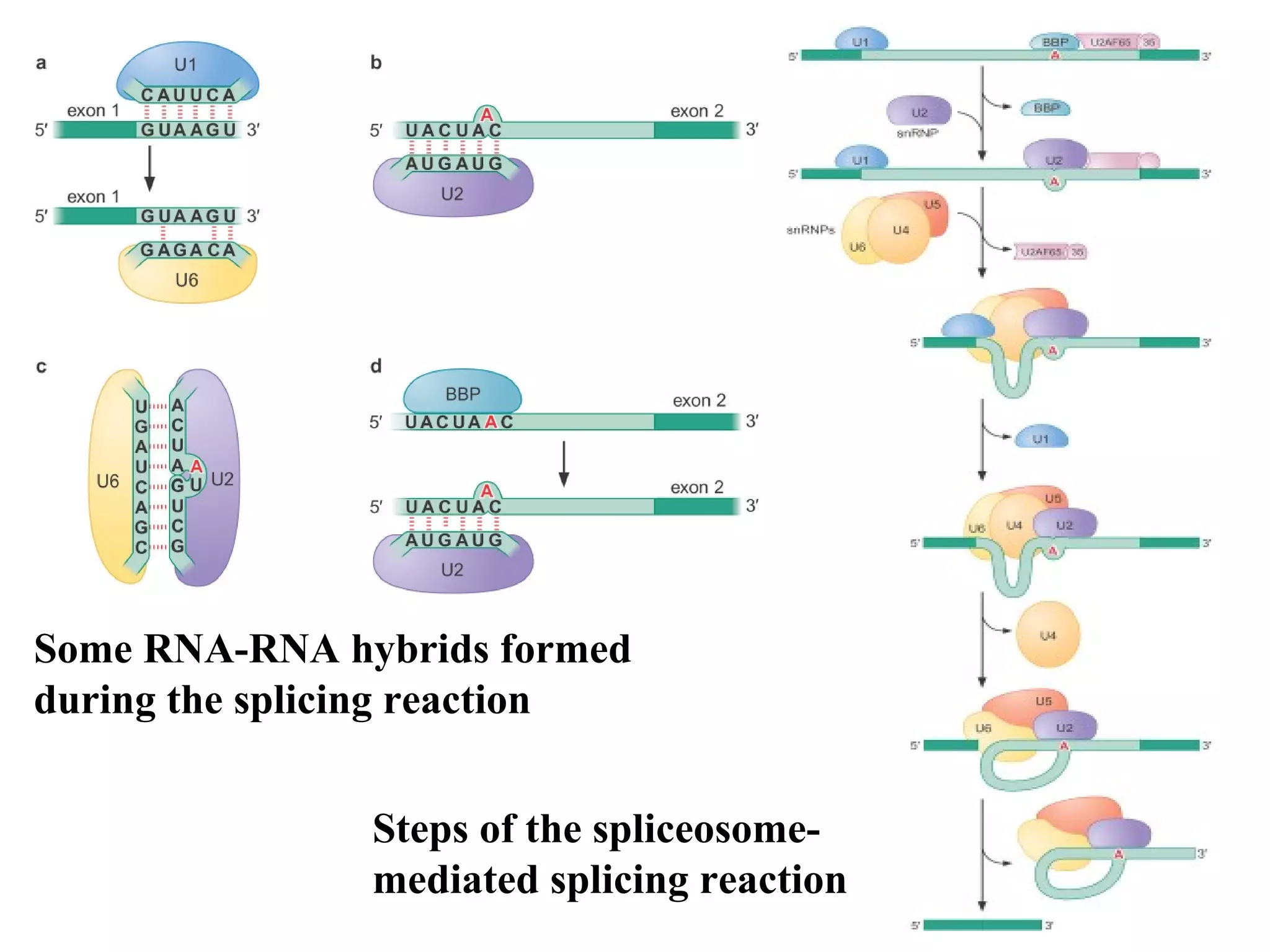
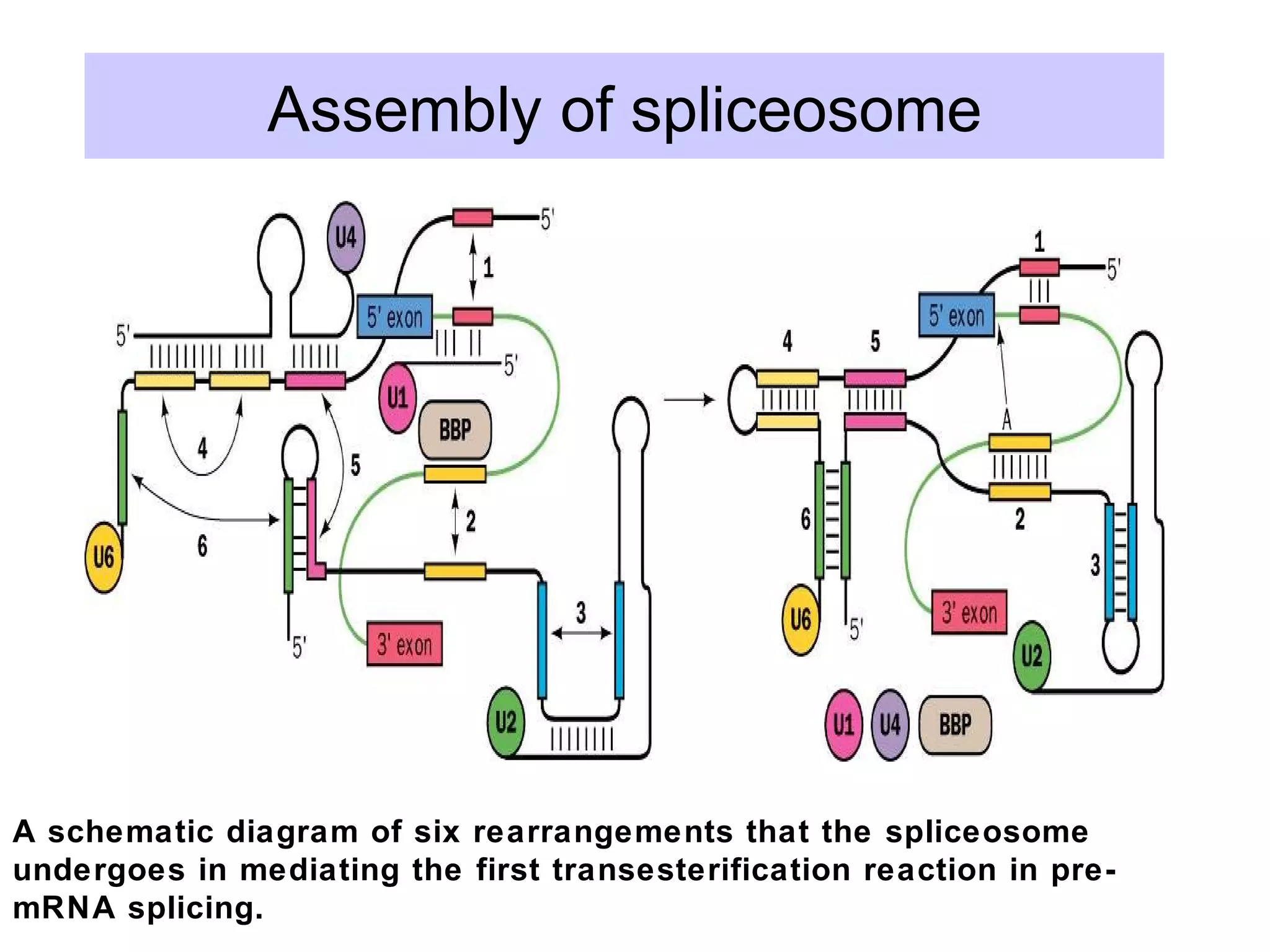
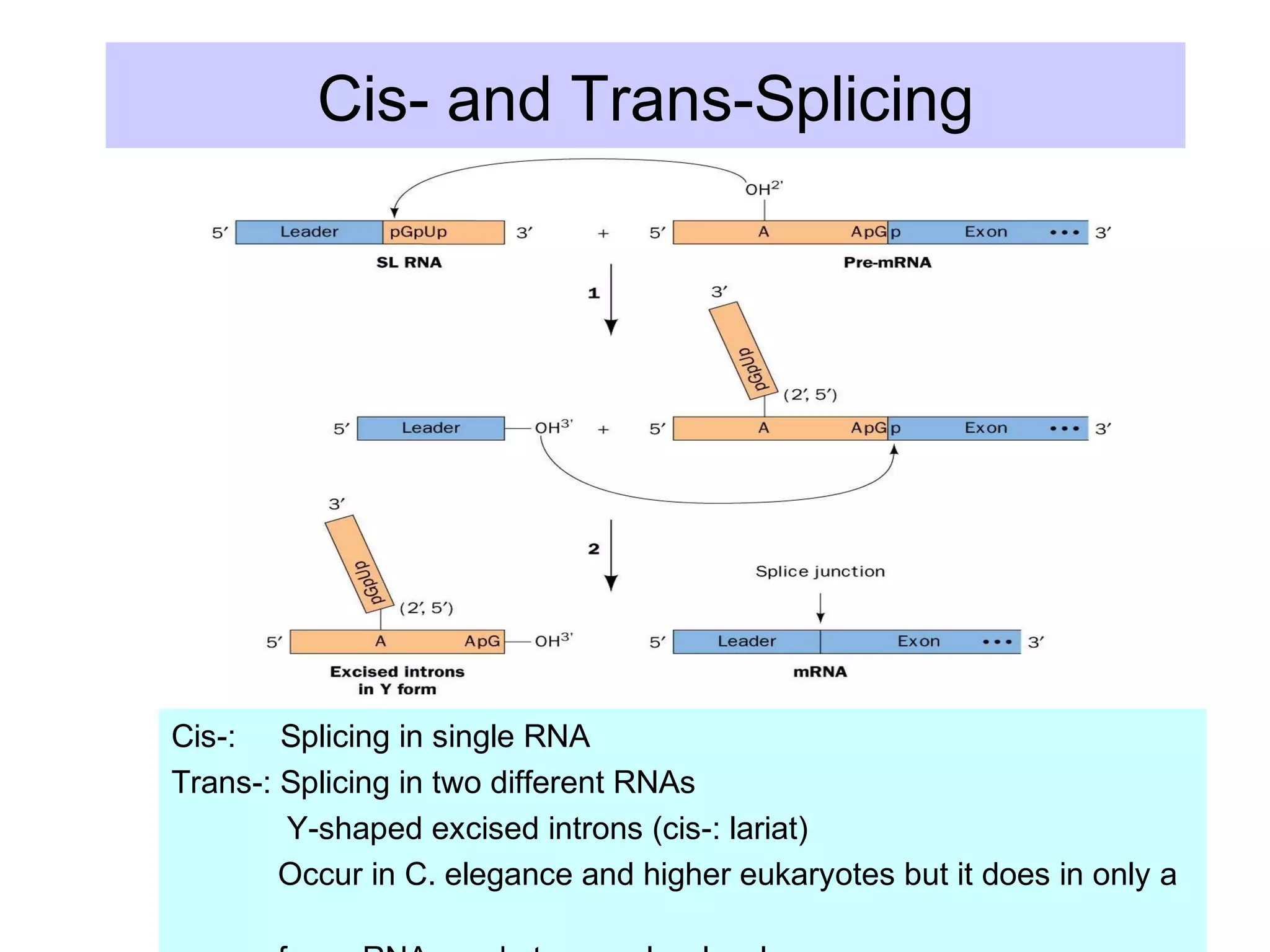
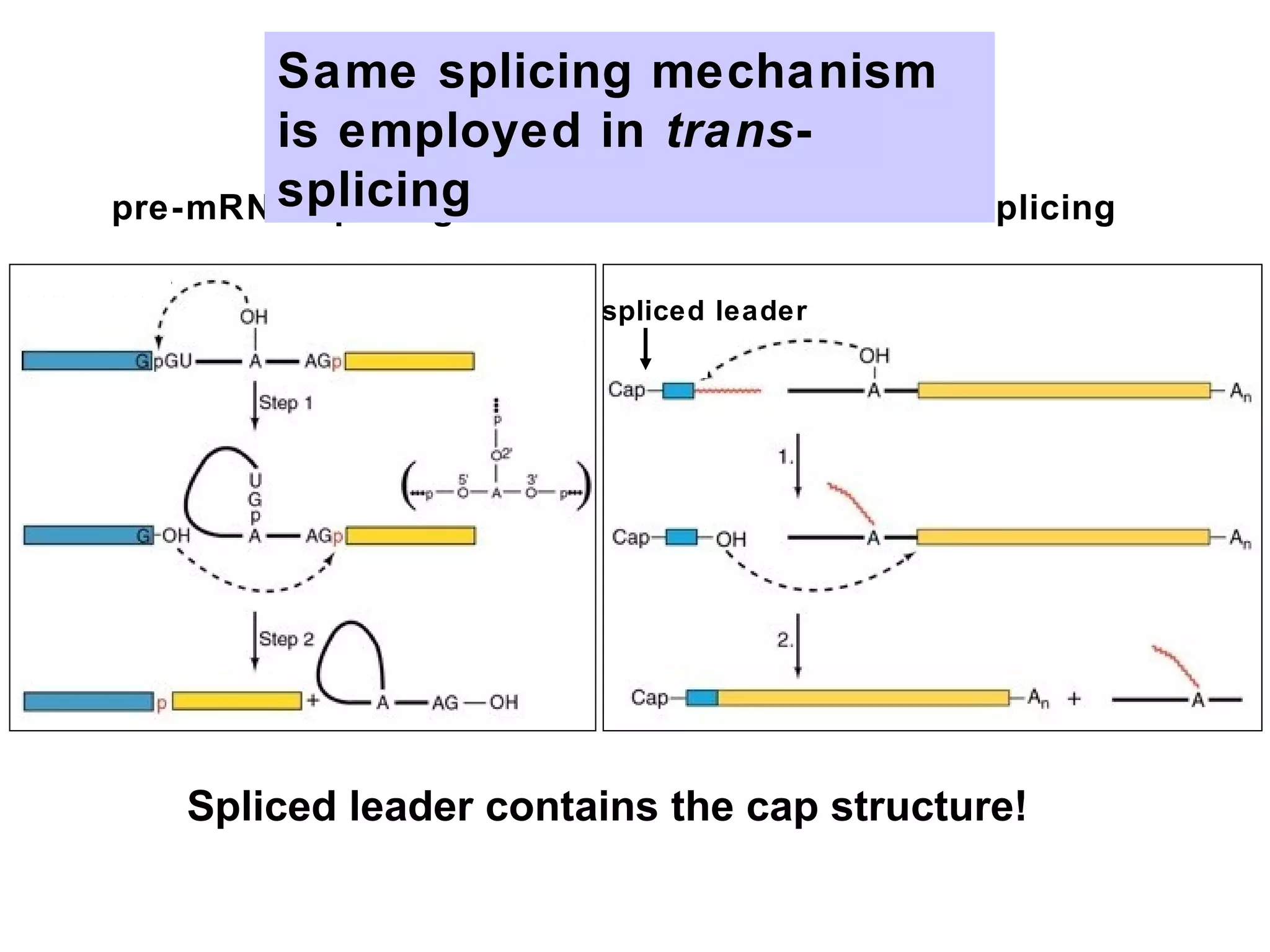
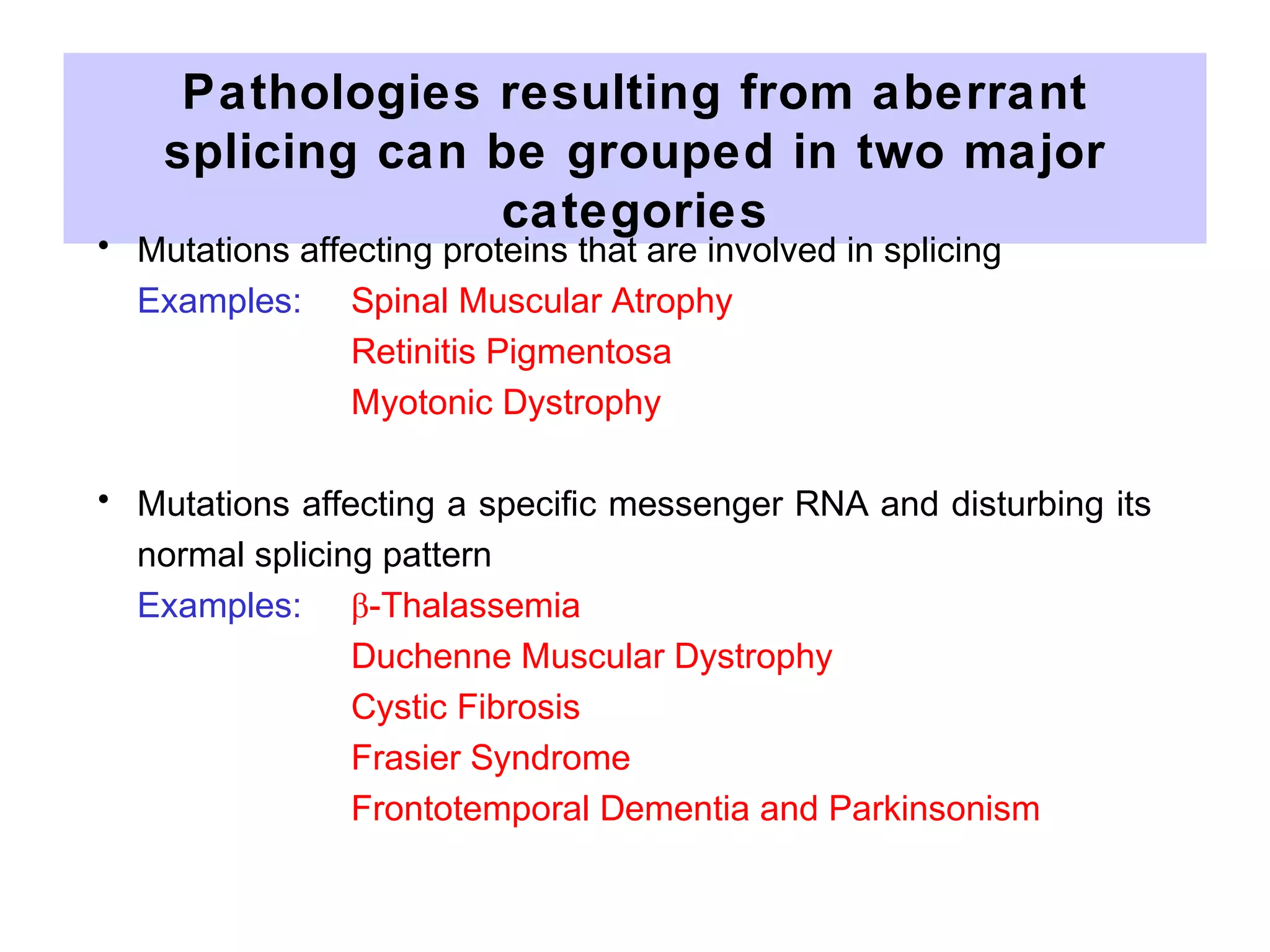
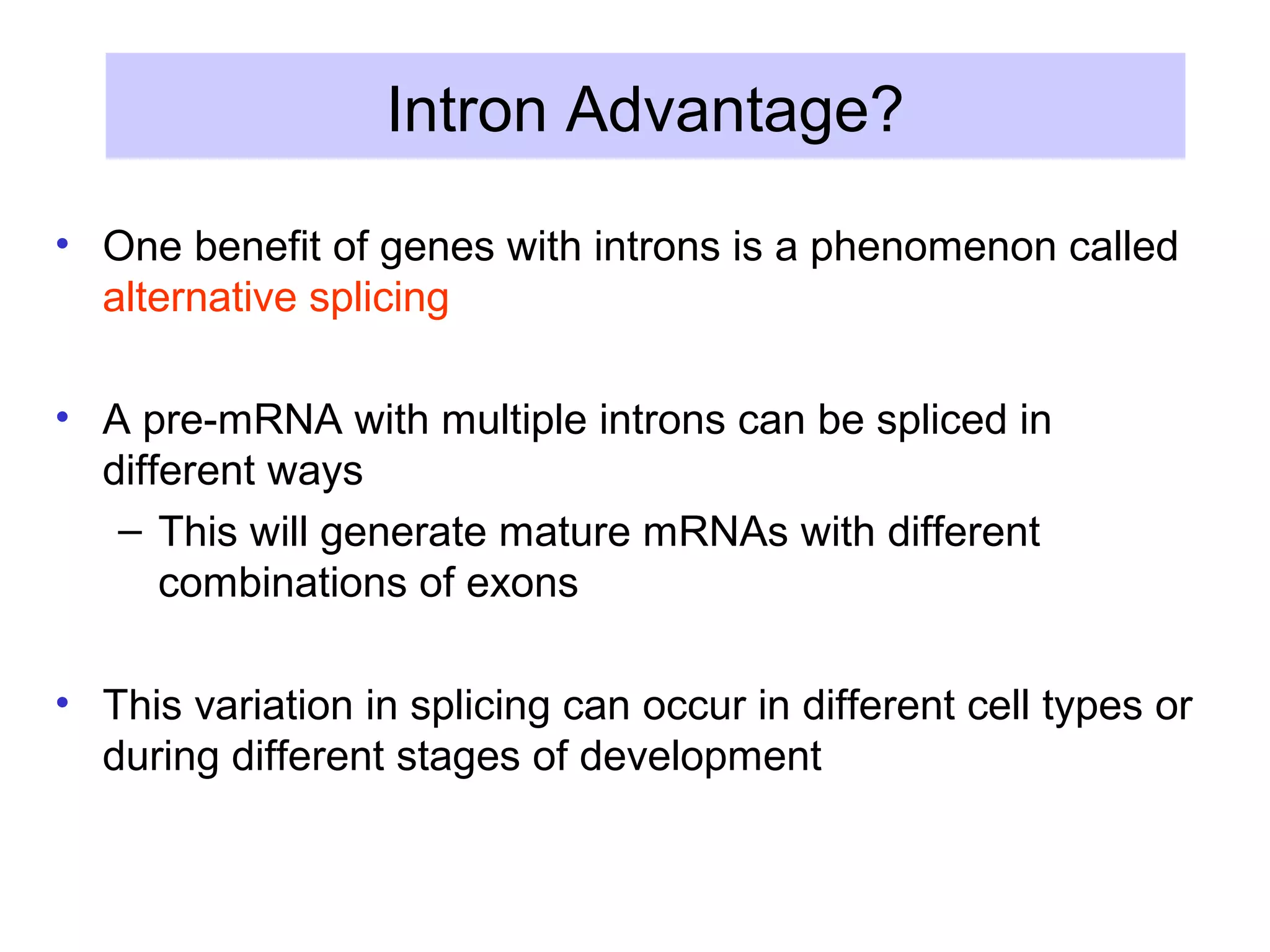
2) Splicing of pre-mRNA involves recognition of splice sites, branch formation using an internal A, and ligation of exons via transesterification reactions. This is catalyzed by the spliceosome, a large complex containing 5 snRNAs and associated proteins.
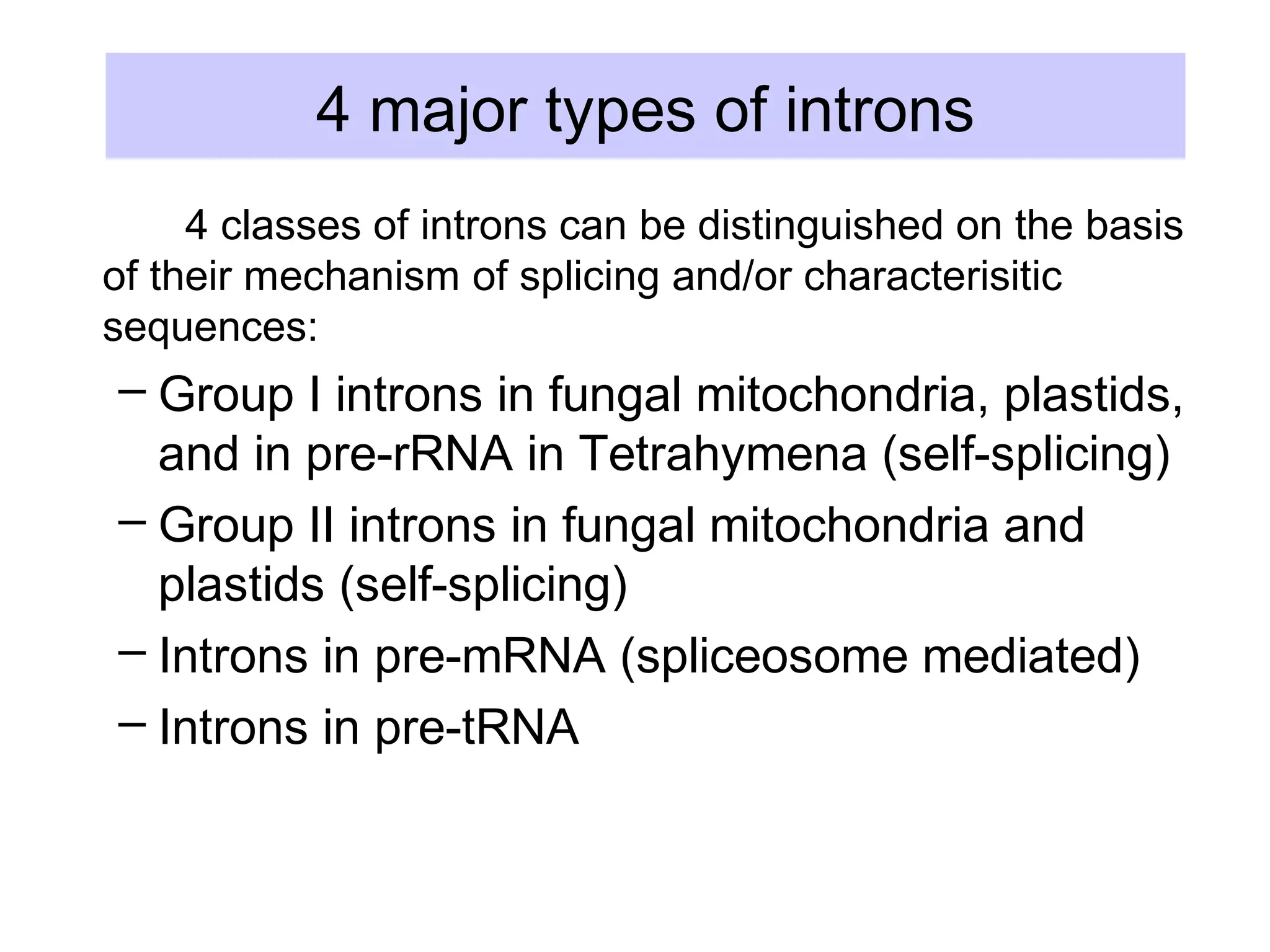
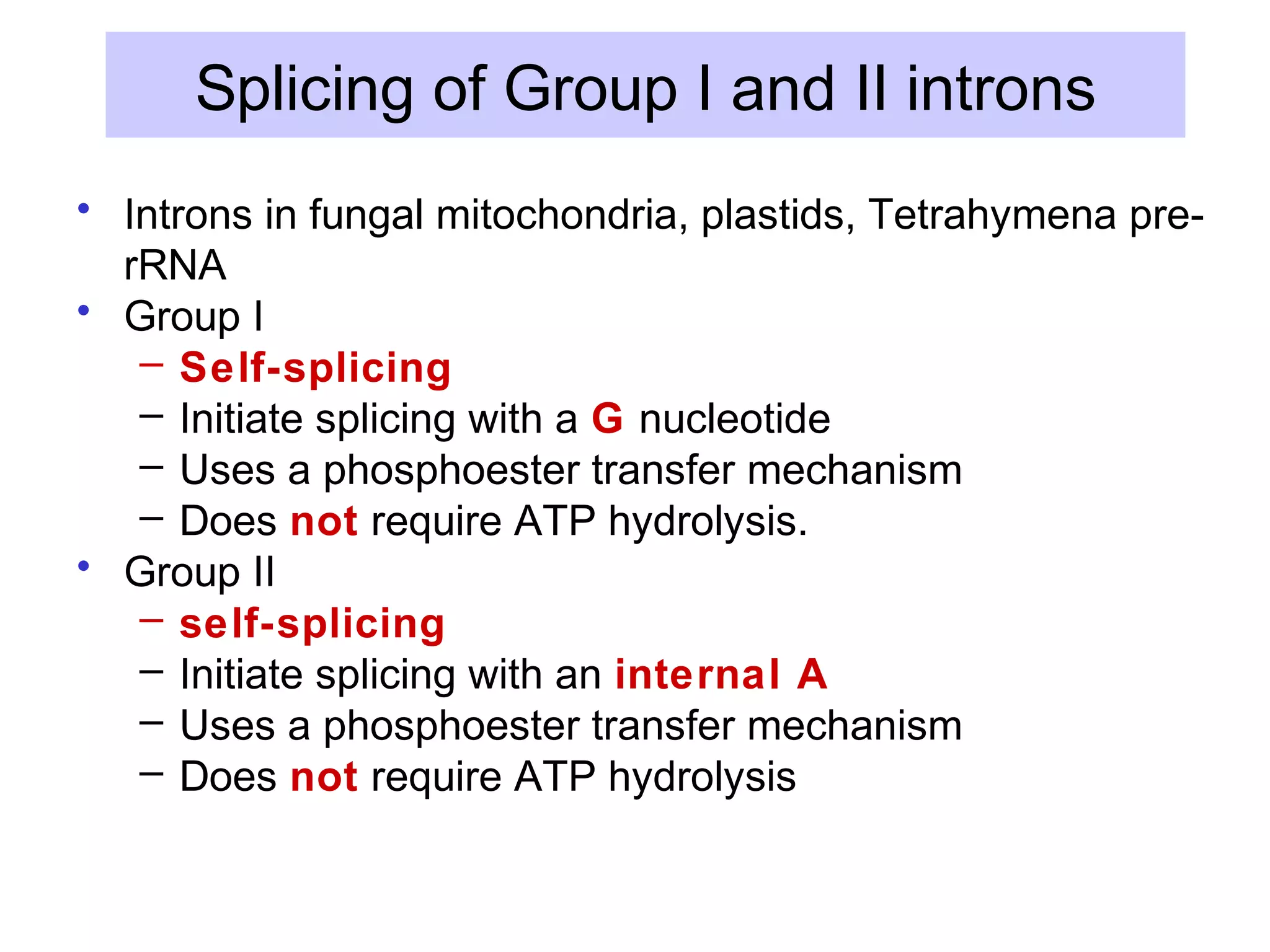
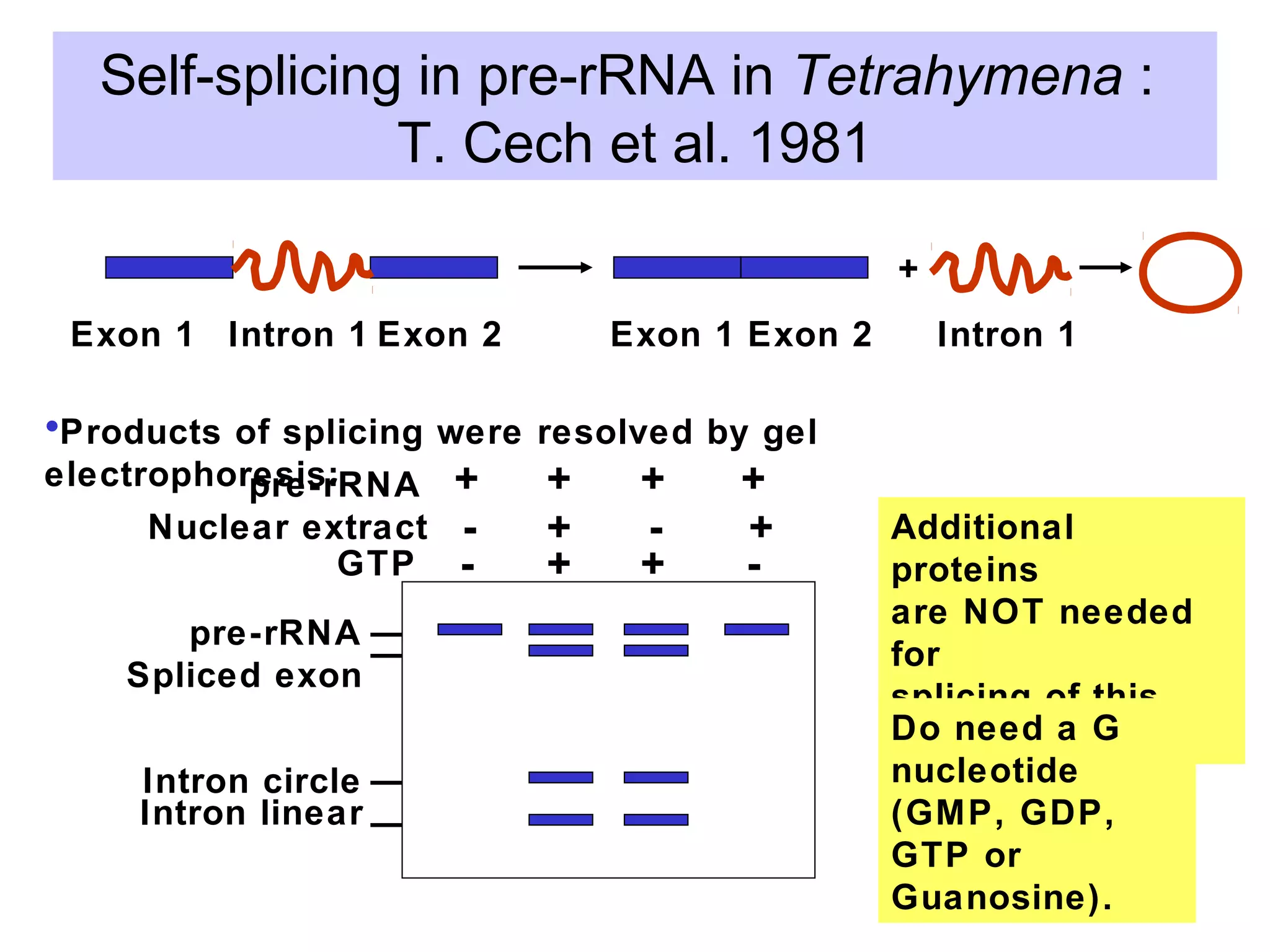
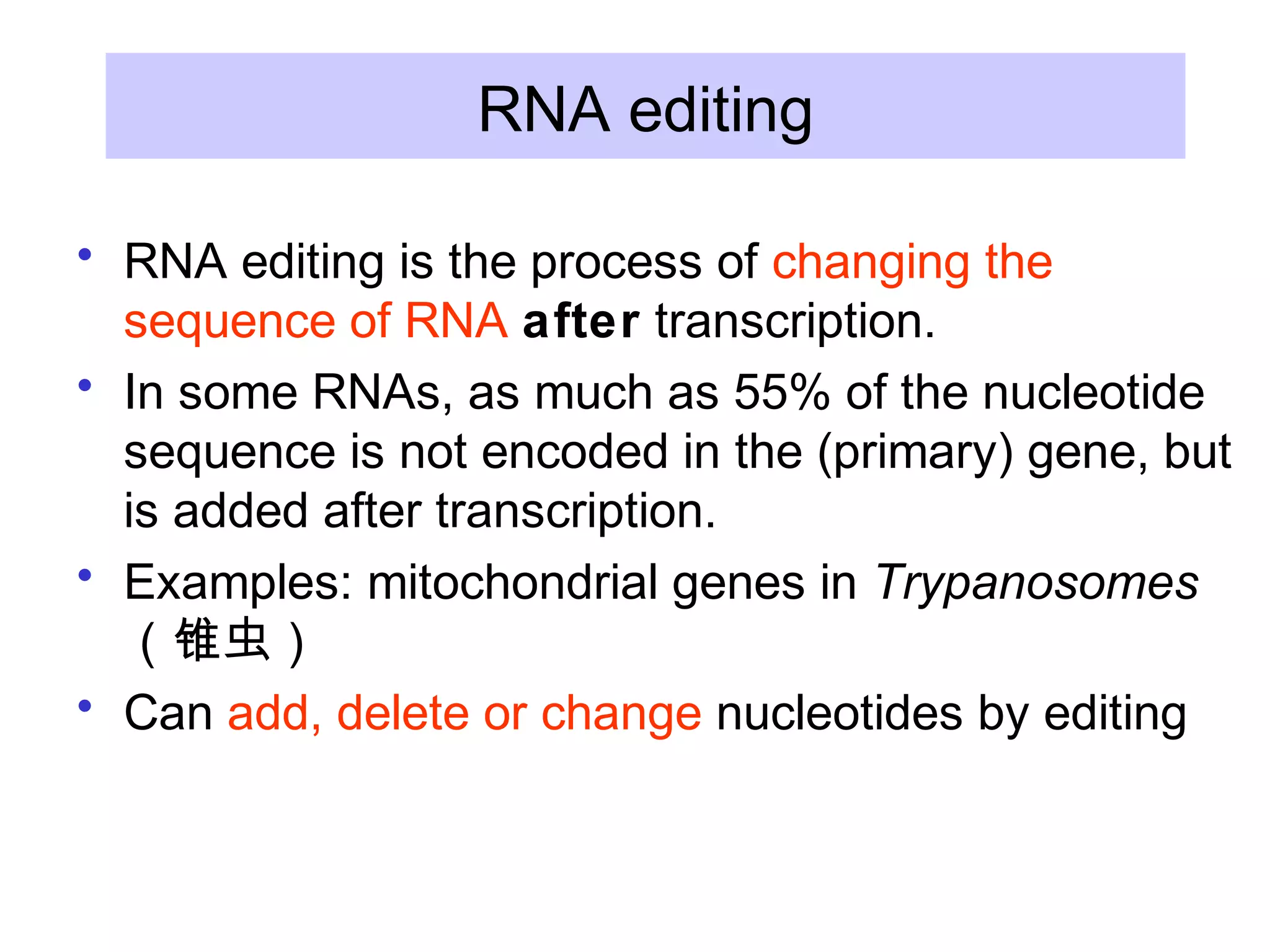
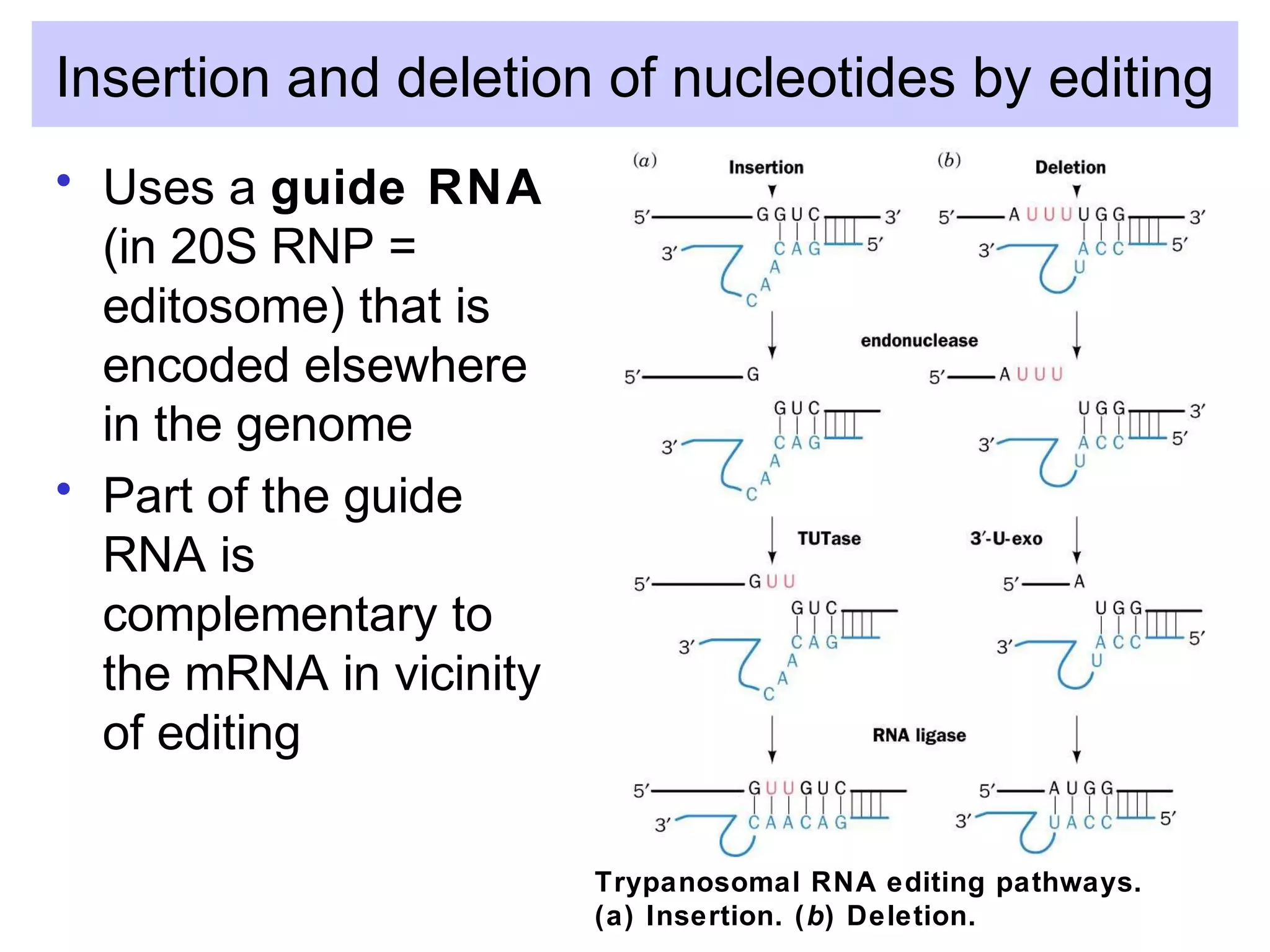
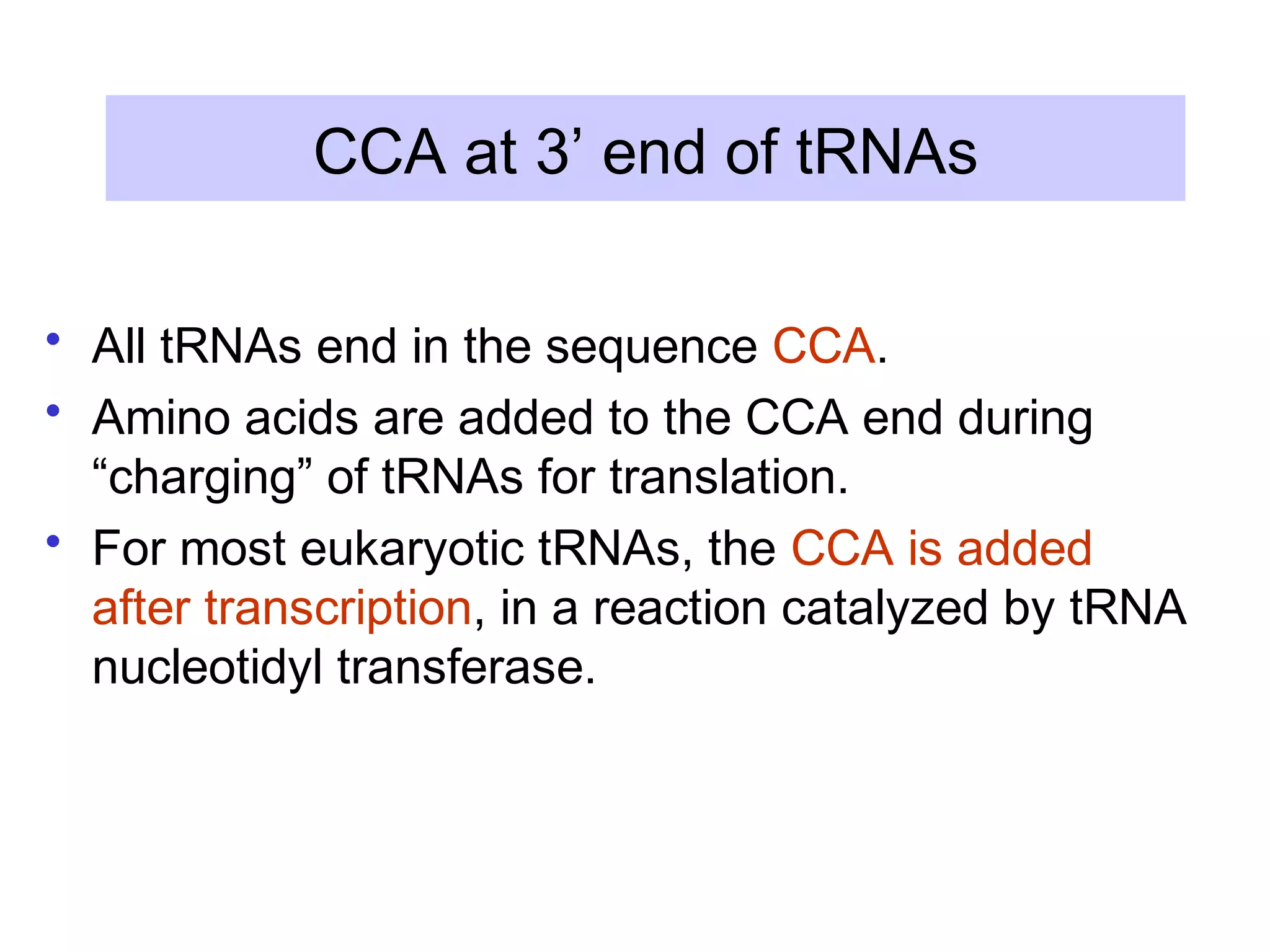
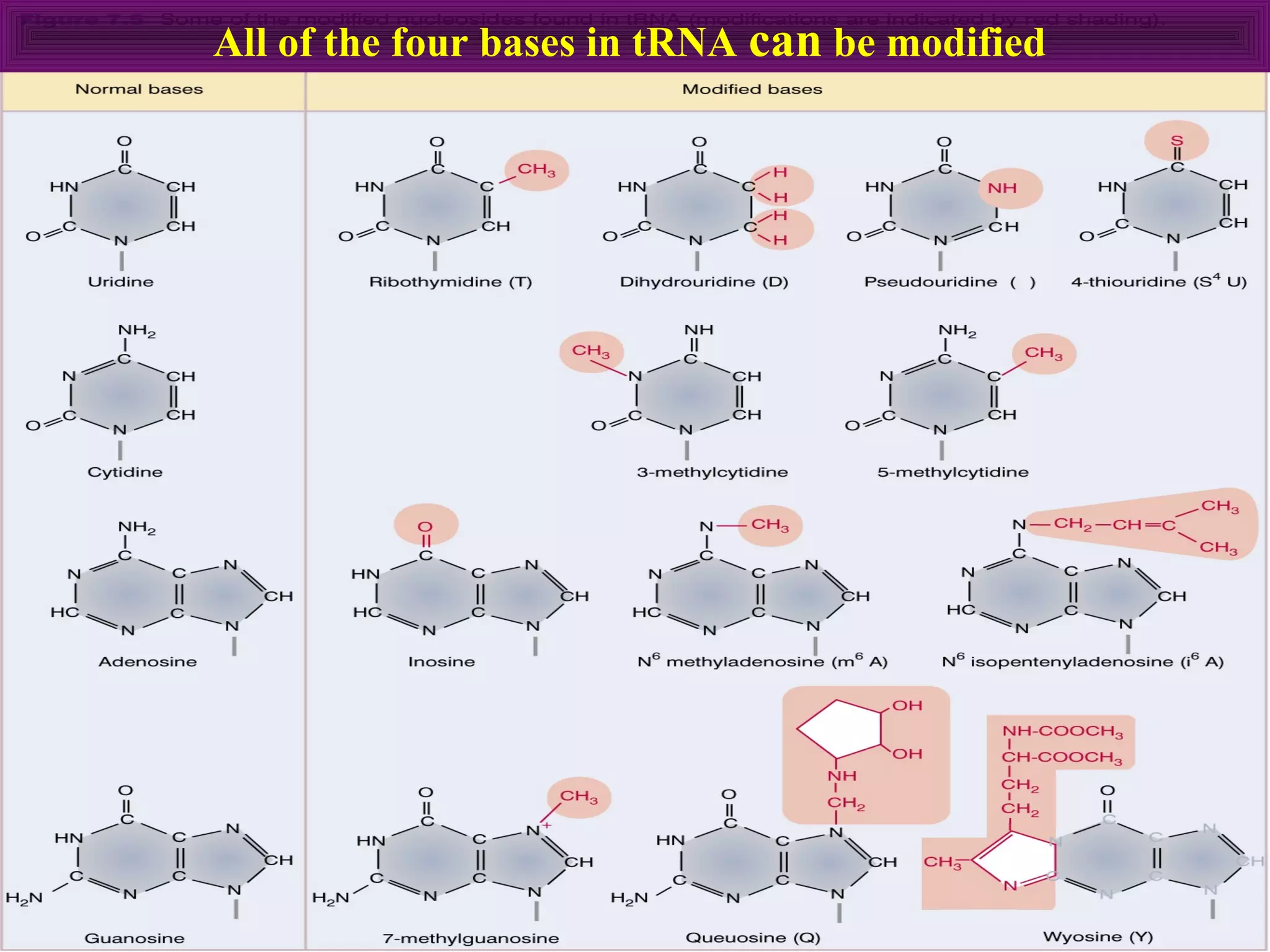
3) Other types of RNA processing addressed include rRNA and tRNA processing, as well as different classes of