This document provides an overview of topics to be covered in a lecture on single neuron models. It will discuss:
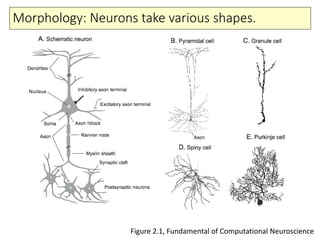
1) The basic anatomy and physiology of neurons including their morphology and membrane properties.
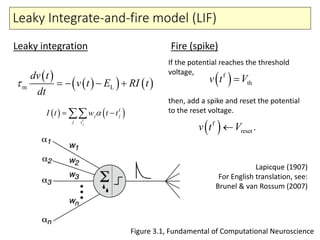
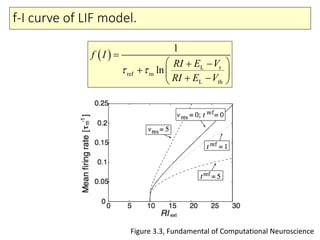
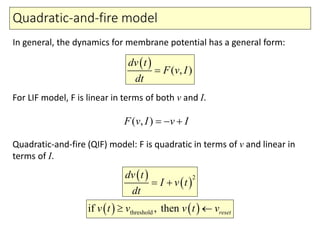
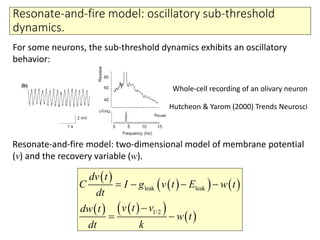
2) Phenomenological models of subthreshold dynamics like the integrate-and-fire, quadratic-and-fire, and resonate-and-fire models.
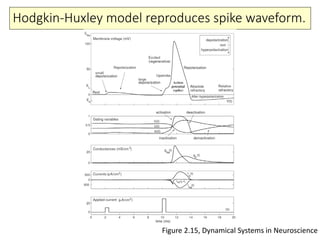
3) Biophysical models of spiking mechanisms including the Hodgkin-Huxley model and its use of ion channels and master equations.
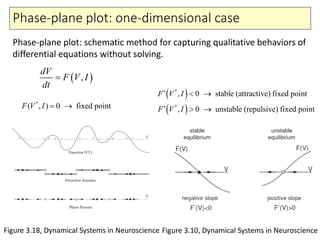
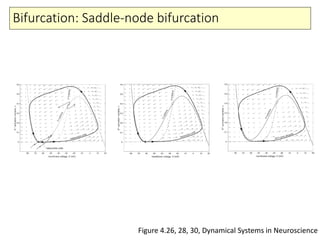
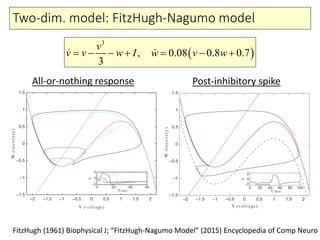
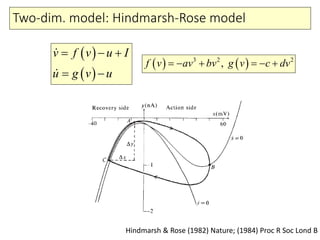
4) Analysis techniques like phase plots and bifurcation analysis applied to models like FitzHugh-Nagumo and Hindmarsh-Rose.
5) Modern single neuron models such









![Ion channels: Nernst equation.
Figure 6.3, Fundamental of Computational Neuroscience
[ ]
[ ]
out
ion in out
in
ion
ln
ion
RT
E E E
zF
≡ − =
EoutEin Nernst equation
[ ]
[ ]
( )
out
out in
in
out
in
ion
ion
zF
E zFRT E E
RT
zF
E
RT
e
e
e
−
− −
−
= =
in[ ] 140mMK+
= out[ ] 3mMK+
=
[ ]
[ ]
out
in
3
ln 61.5ln 102mV
140
K
KRT
E
F K
= = = −
Potassium ion](https://image.slidesharecdn.com/ss201601singleneurons-160830235901/85/JAIST-2016-01-Single-neuron-models-16-320.jpg)