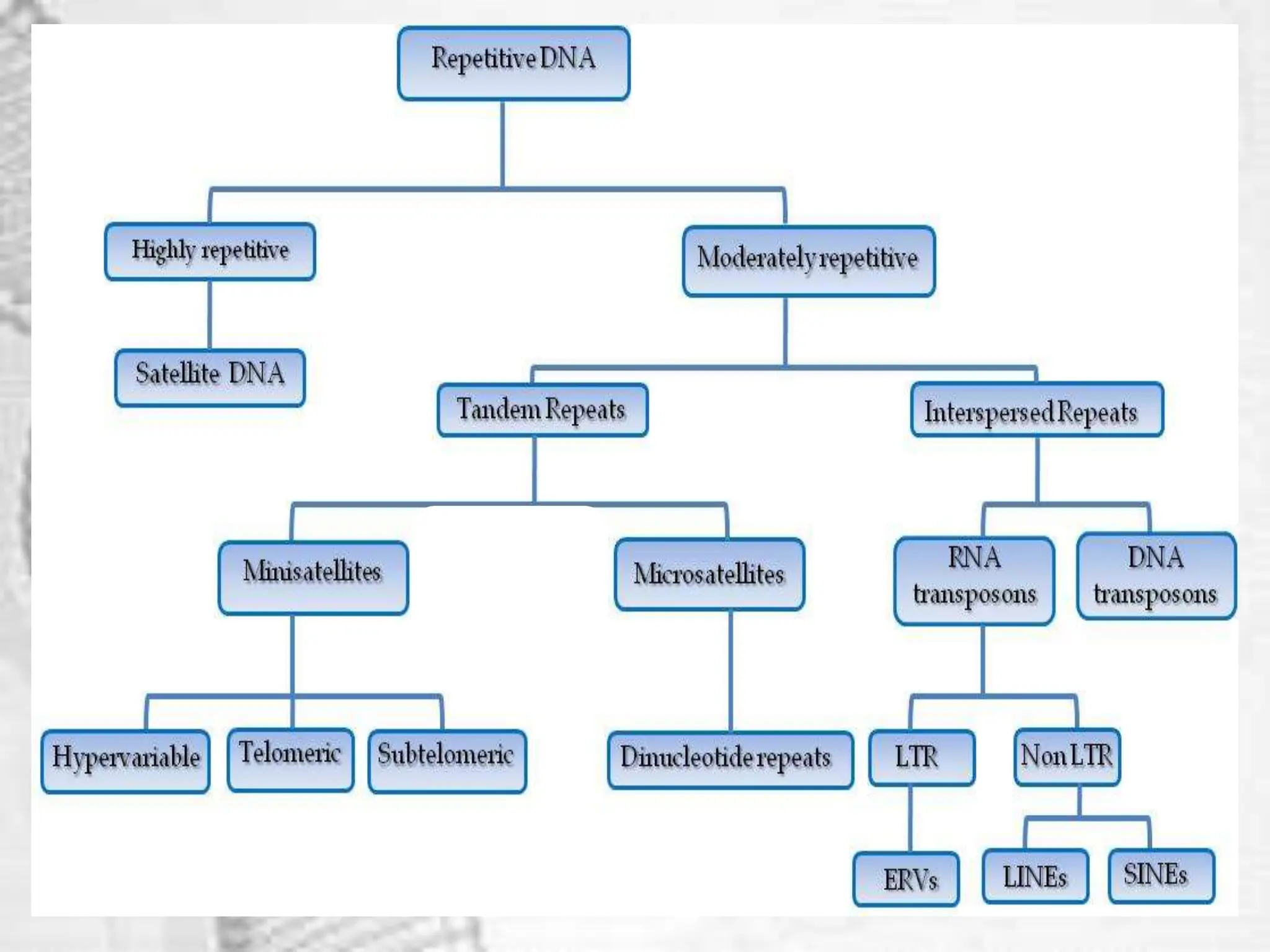
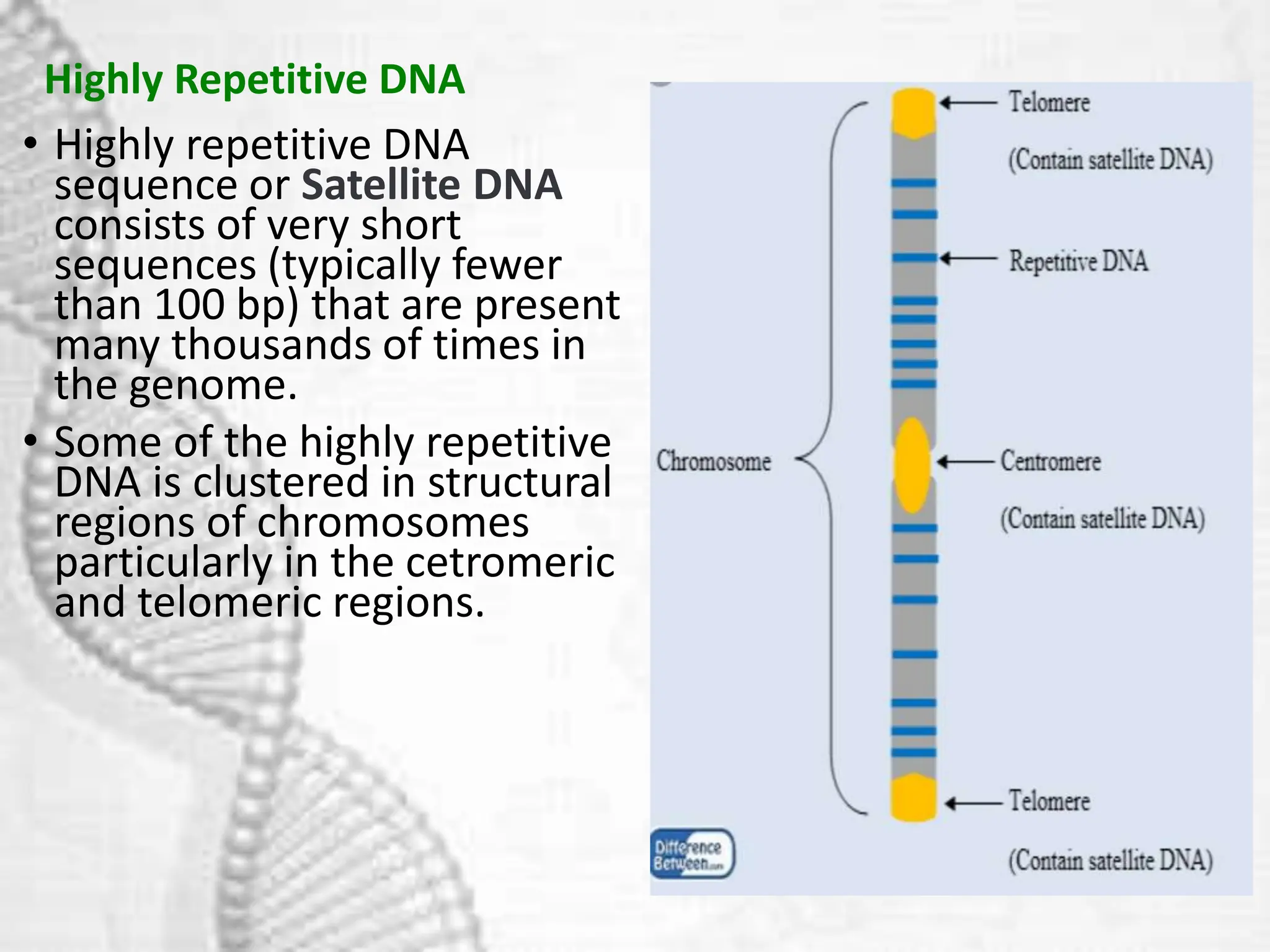
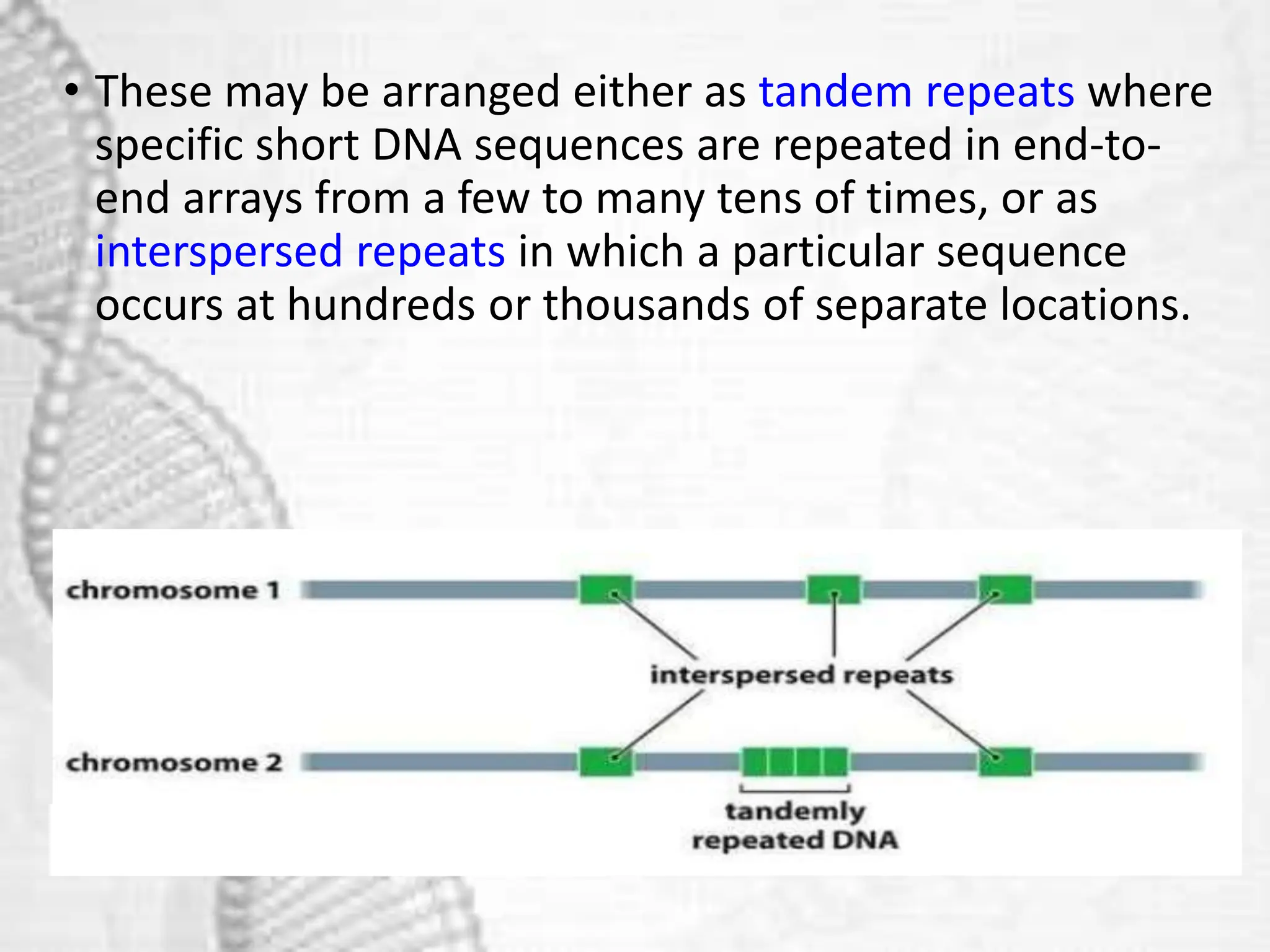
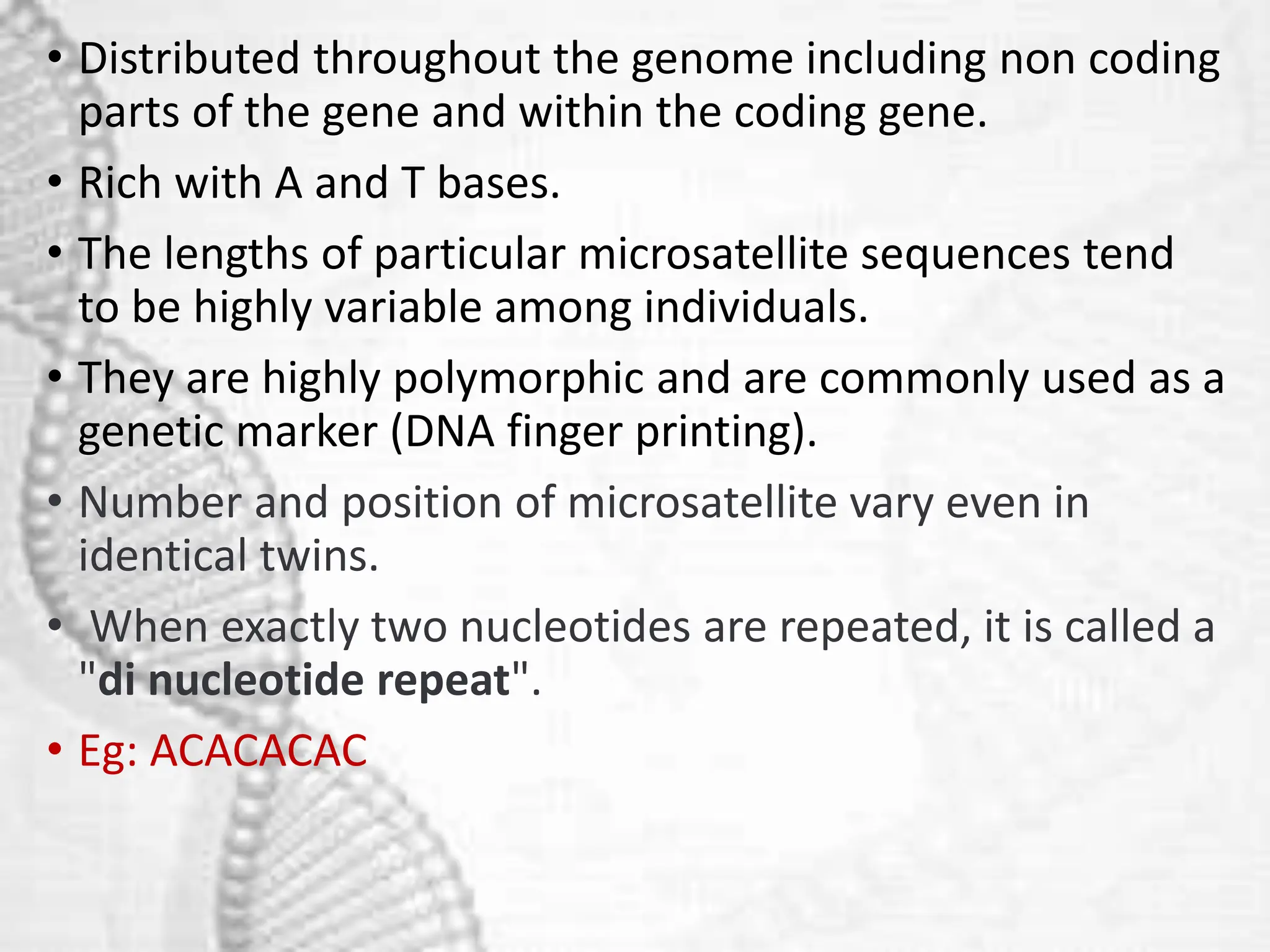
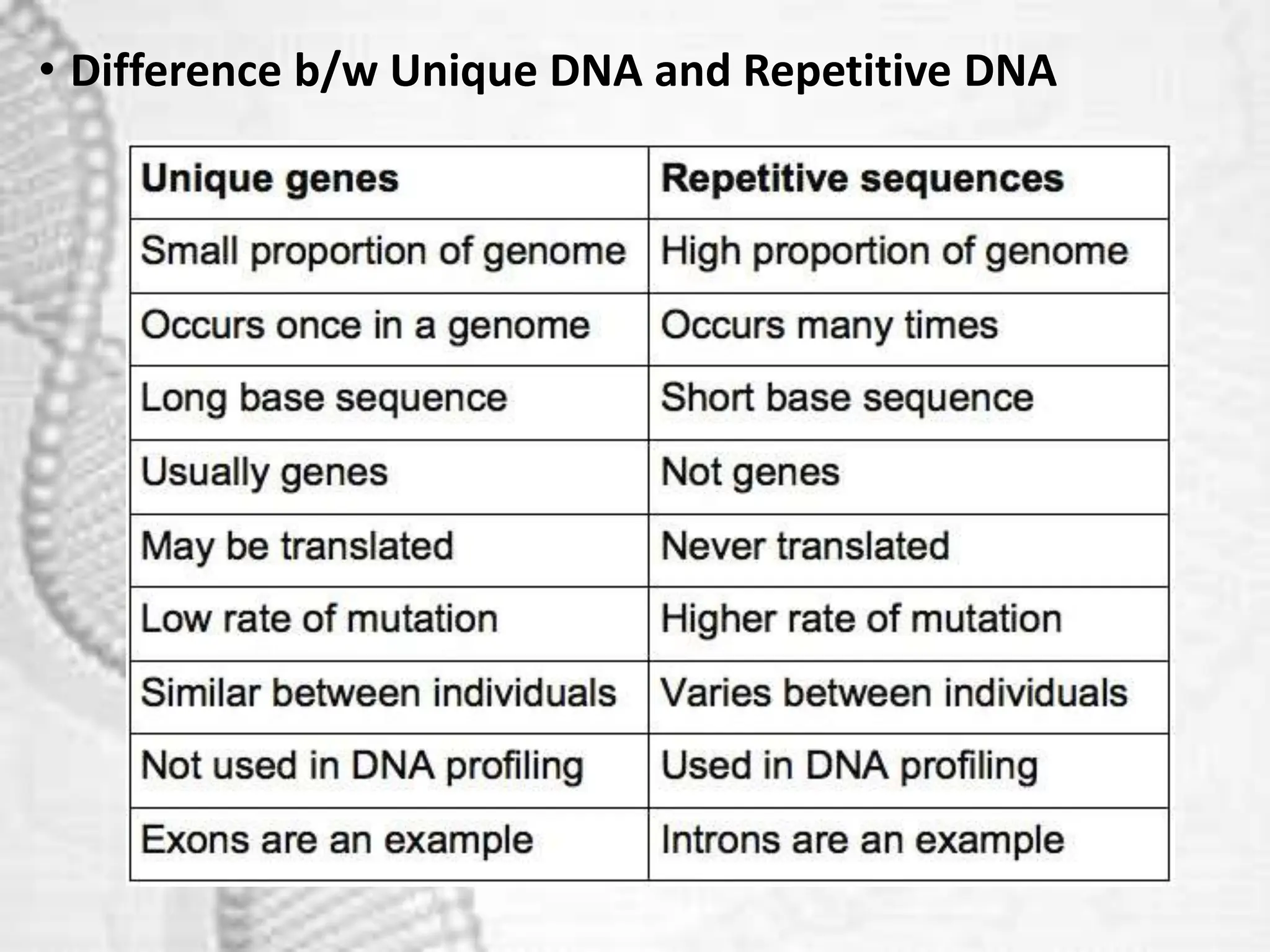
The document discusses unique and repetitive DNA sequences found in eukaryotic genomes. It defines unique DNA as sequences present in a single copy that encode for proteins. Repetitive DNA makes up a large portion of eukaryotic genomes and includes highly repetitive sequences like satellite DNA near centromeres, and moderately repetitive sequences dispersed throughout genomes like transposons. The repetitive elements are further classified based on length and copy number as tandem repeats including satellites, minisatellites and microsatellites.