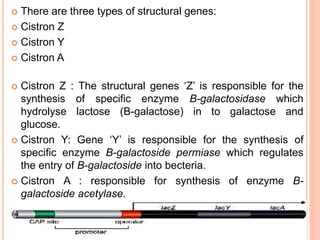
The document discusses gene regulation mechanisms, focusing on the operon model first proposed by Jacob and Monod in 1961, which describes the processes of induction and repression in prokaryotes like E. coli. It explains the structural components of the operon, including structural and controller genes, and illustrates examples such as the lac and trp operons, detailing their regulatory mechanisms. Ultimately, it emphasizes the importance of gene regulatory proteins in controlling transcription and the expression of genes in prokaryotic cells.