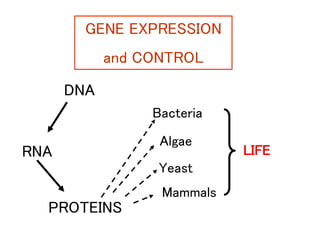
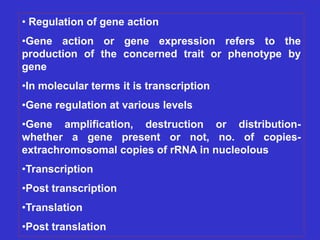
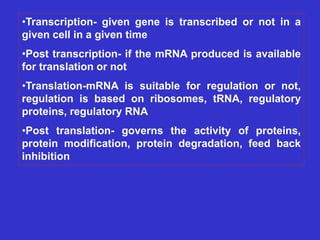
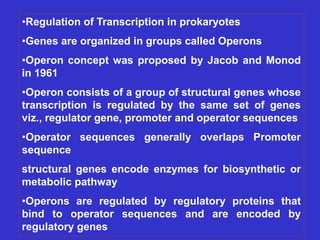
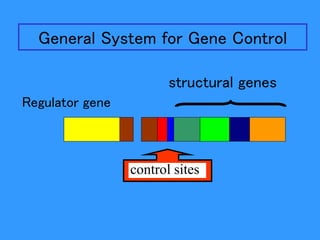
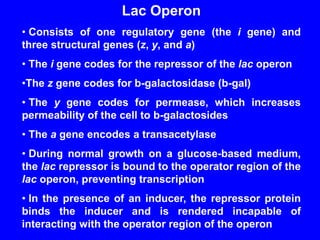
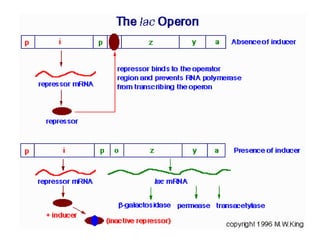
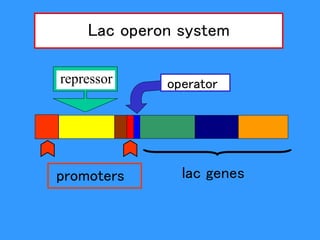
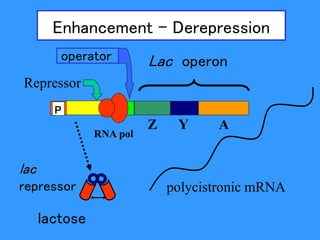
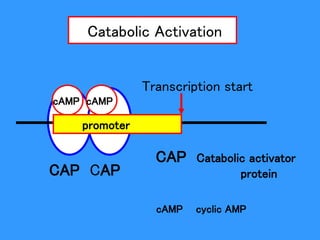
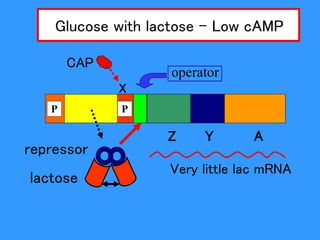
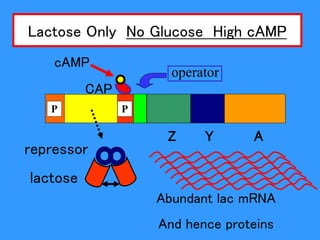
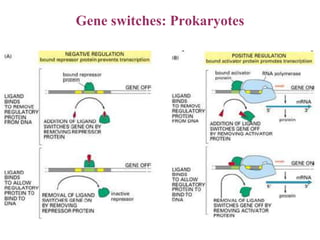
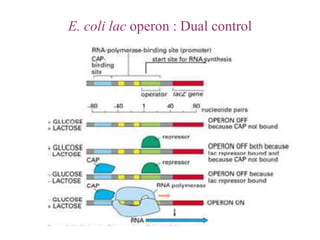
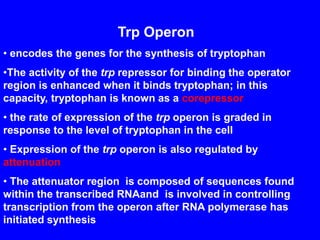
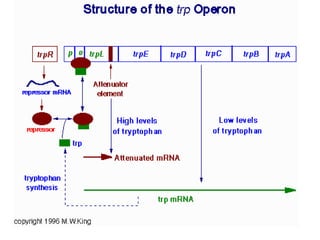
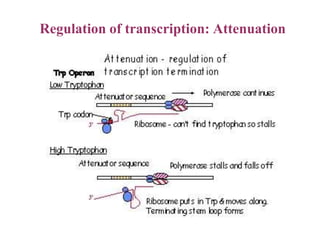
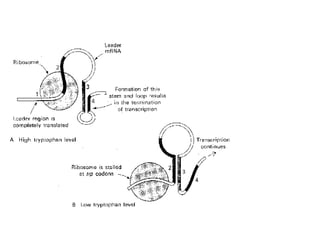
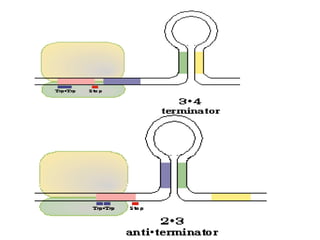
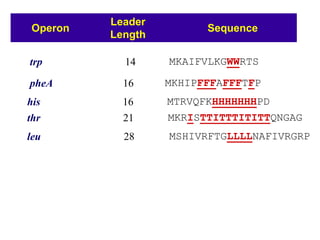
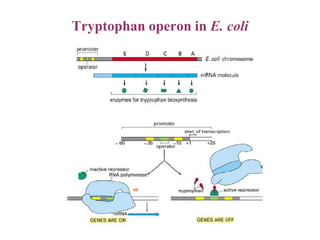
Gene expression in bacteria is often regulated through operons. The lac operon in E. coli consists of structural genes that encode proteins for lactose metabolism regulated by the lac repressor. In the presence of lactose and absence of glucose, cAMP levels rise and activate the catabolite activator protein (CAP), allowing transcription. The trp operon encodes tryptophan synthesis and is regulated by the trp repressor as well as attenuation, where ribosome movement influences RNA structure and termination. Operons demonstrate how prokaryotes efficiently coordinate gene expression through transcriptional control mechanisms.