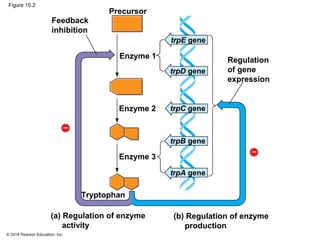
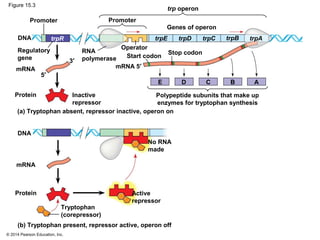
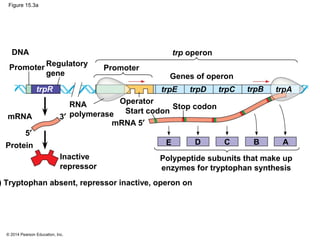
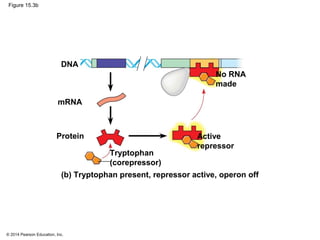
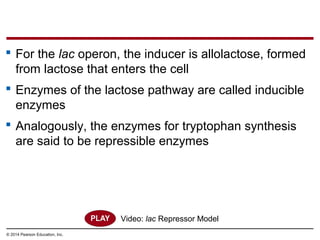
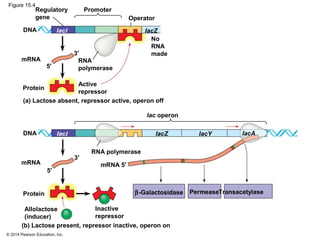
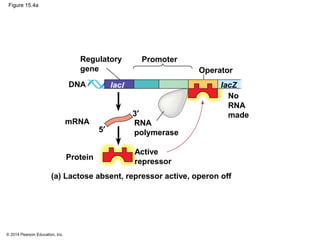
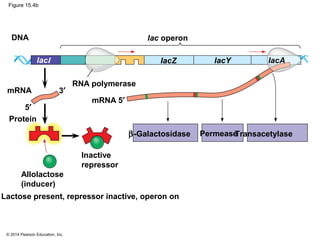
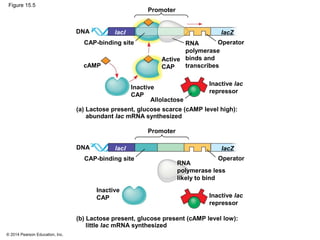
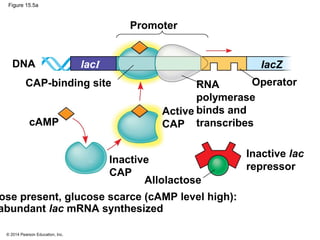
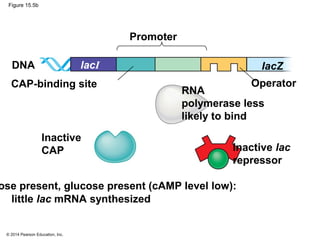
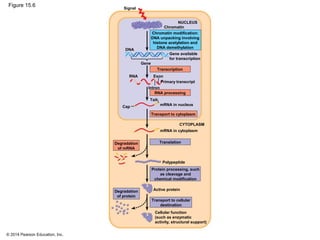
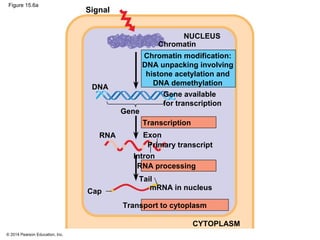
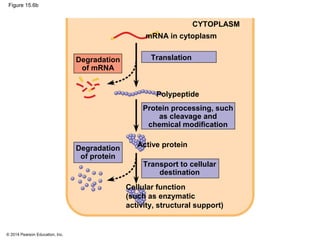
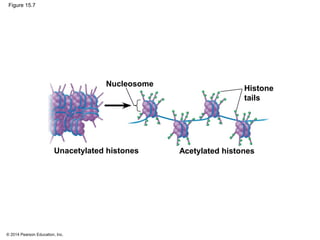
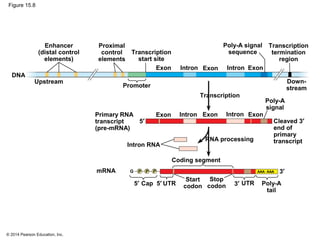
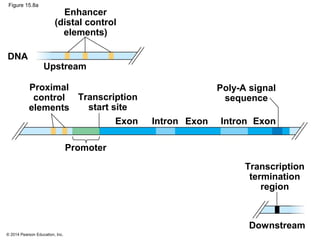
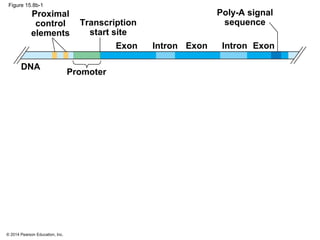
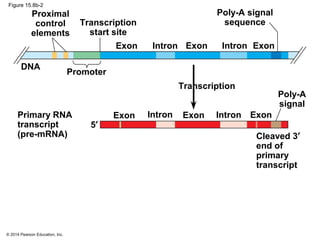
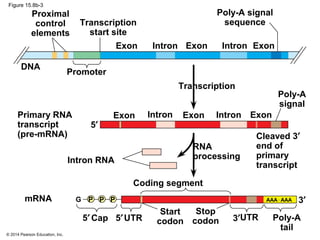
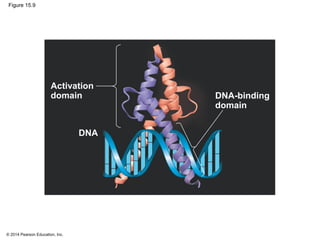
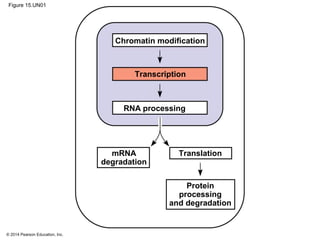
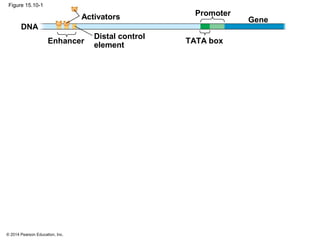
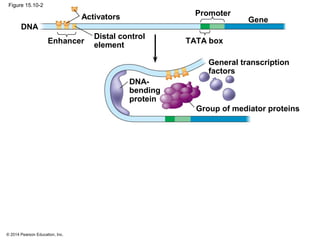
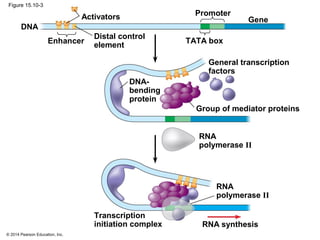
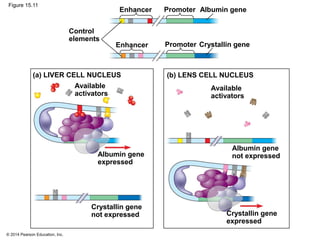
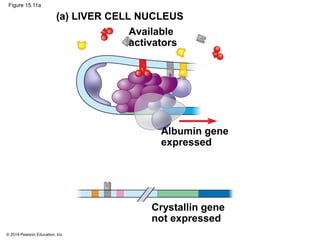
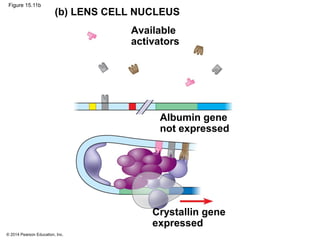
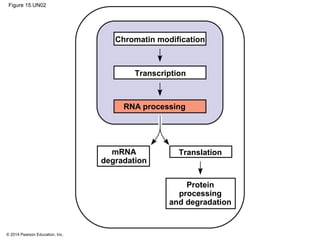
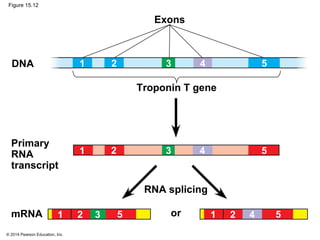
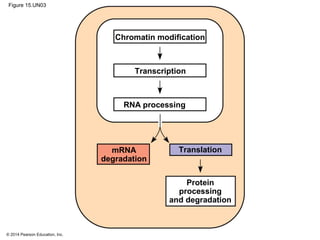
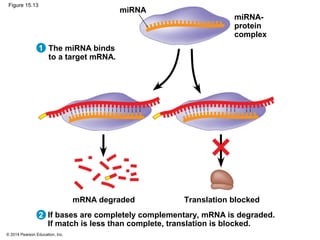
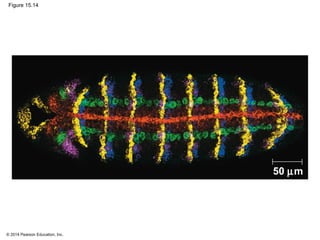
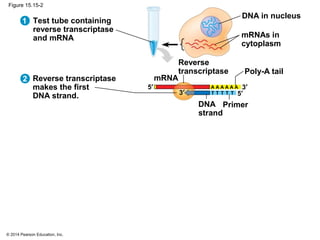
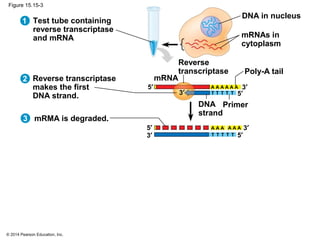
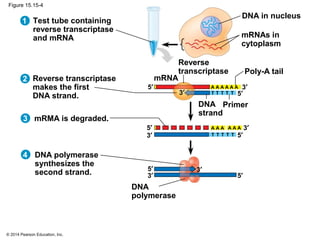
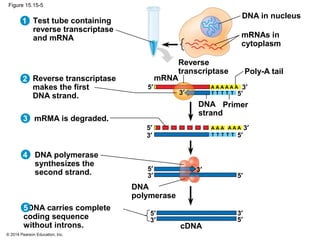
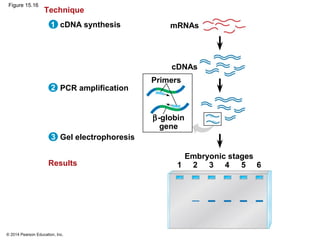
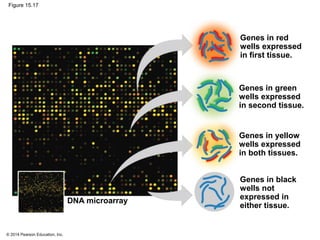
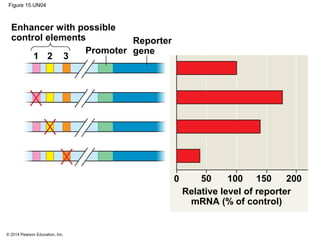
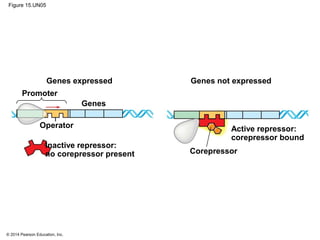
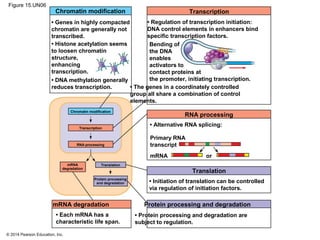
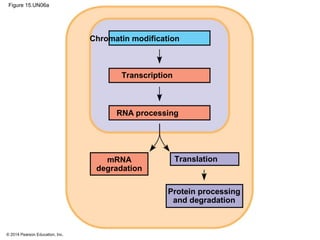
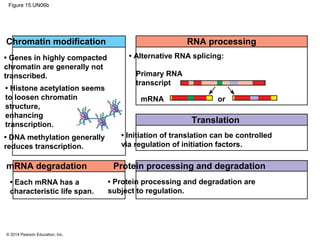
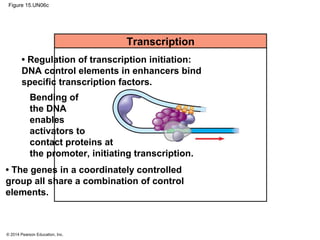
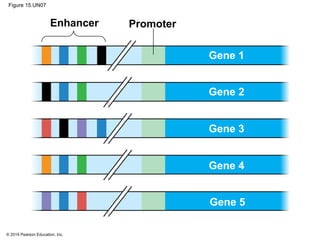
Bacteria and eukaryotes regulate gene expression in response to environmental changes. In bacteria, groups of related genes are often controlled by a single "on-off switch" called an operon. The lac and trp operons in E. coli are examples of negative gene regulation where a repressor protein binds to DNA and blocks transcription. Eukaryotes regulate gene expression at many additional stages including chromatin structure, histone modifications, transcription initiation, RNA processing, and translation. Differential gene expression allows cells with identical genomes to develop specialized structures and functions.