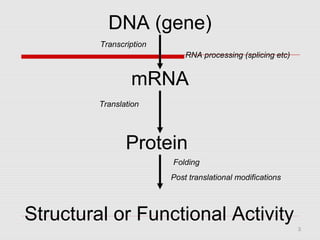
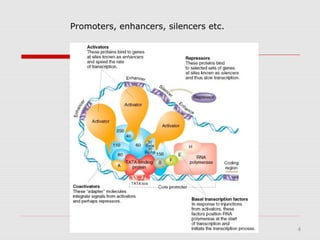
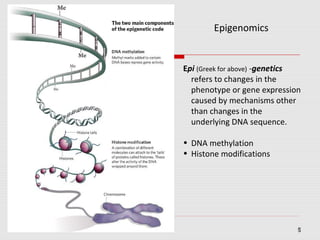
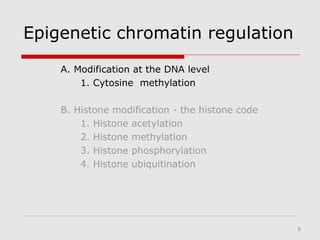
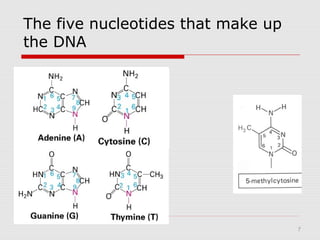
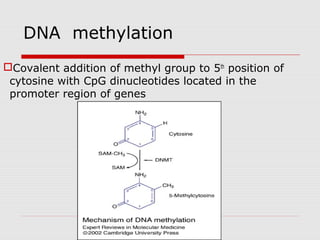
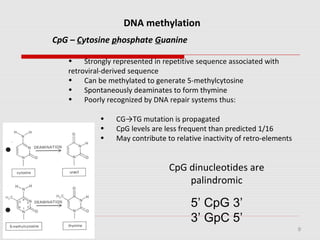
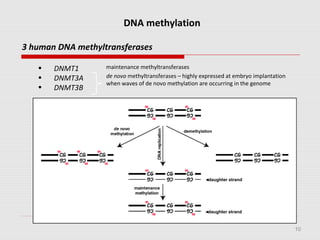
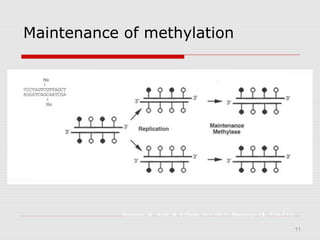
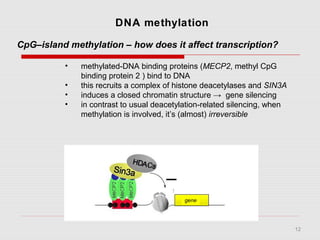
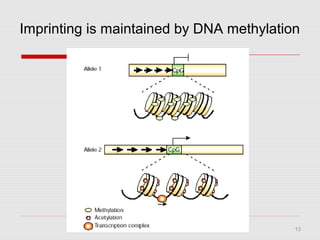
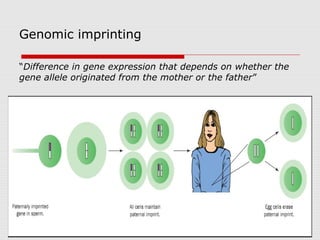
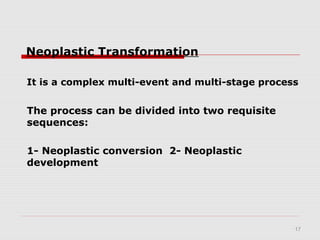
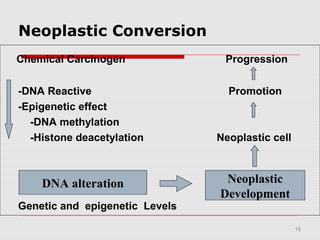
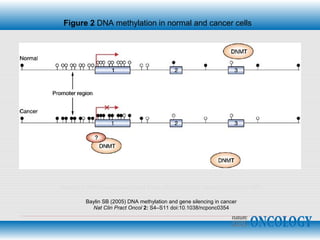
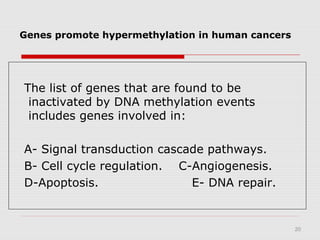
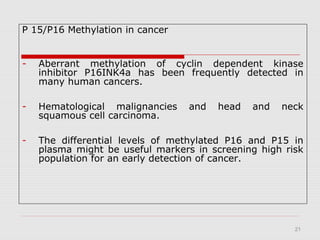
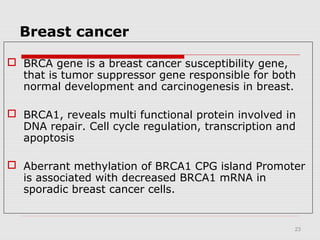
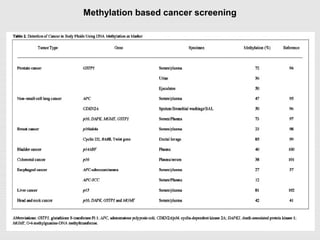
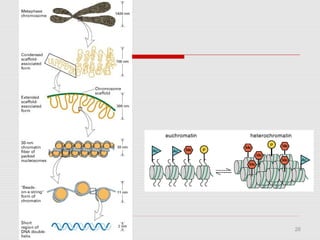
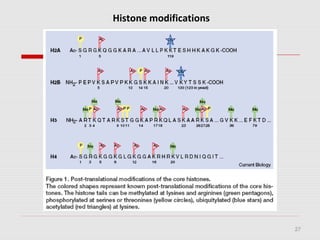
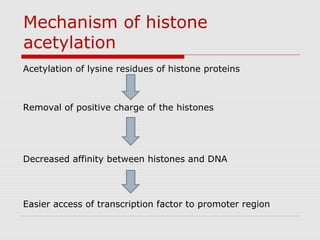
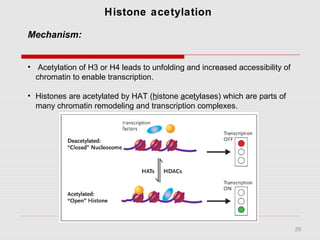
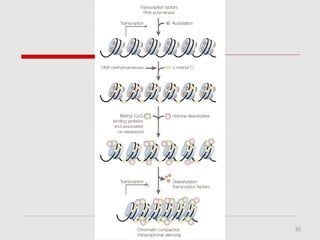
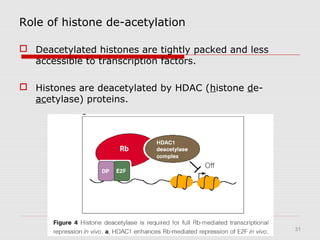
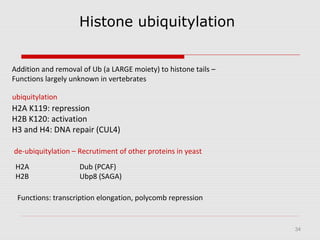
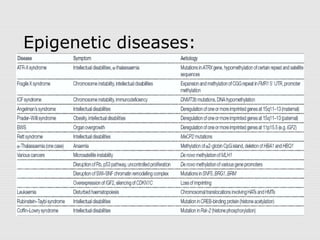
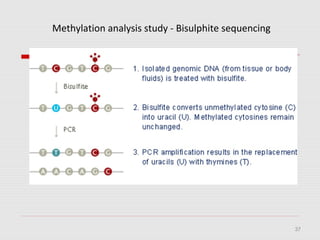
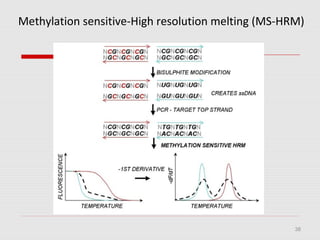
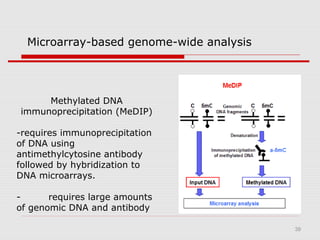
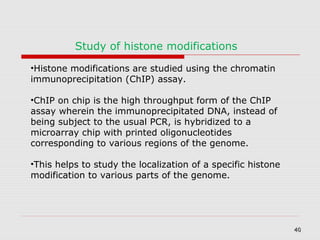
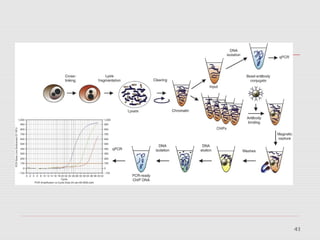
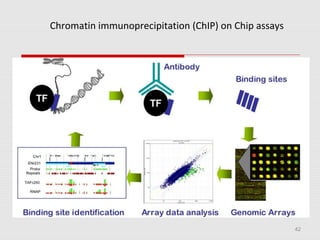
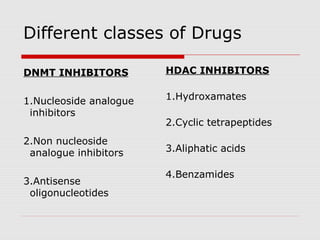
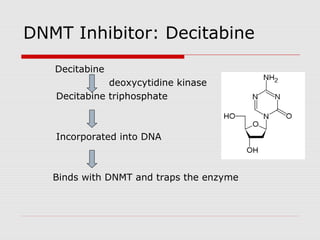
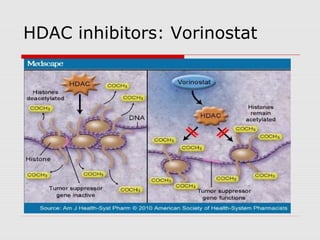
The document discusses epigenetics and the epigenome. It describes the key components of the epigenetic code, including DNA methylation and various histone modifications. It explains how these modifications can regulate gene expression and affect processes like transcription, development, and disease states like cancer. The document also outlines several methods for studying the epigenome, such as bisulfite sequencing and chromatin immunoprecipitation assays. Finally, it discusses potential therapeutic approaches that target the epigenome, including drugs that inhibit DNA methyltransferases and histone deacetylases.