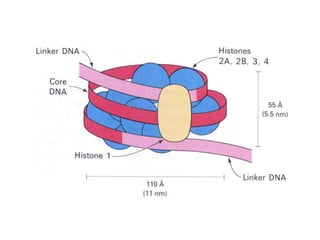
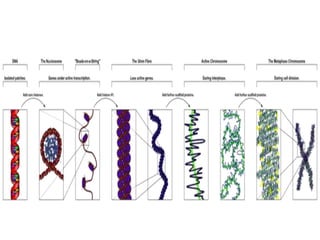
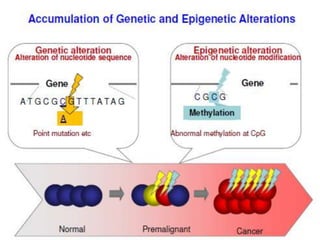
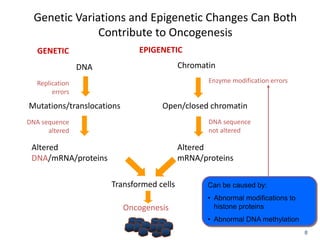
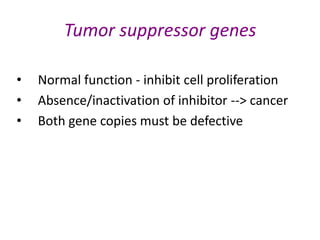
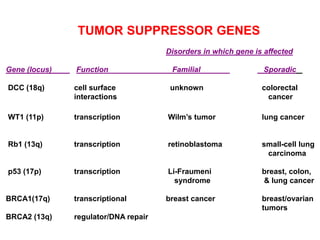
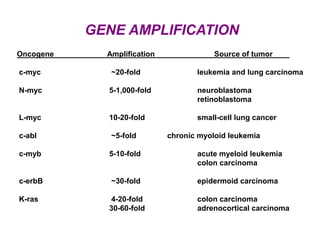
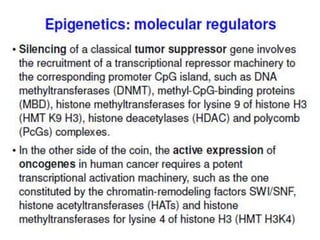
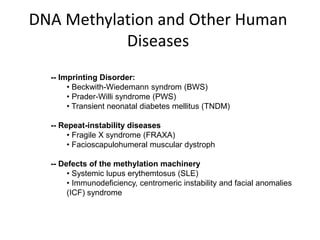
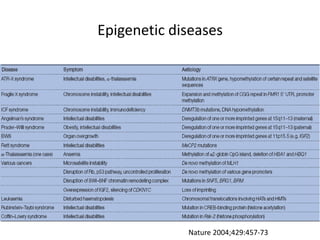
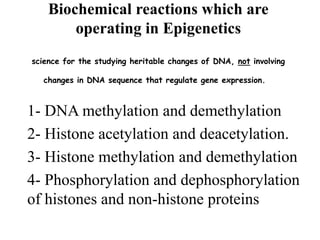
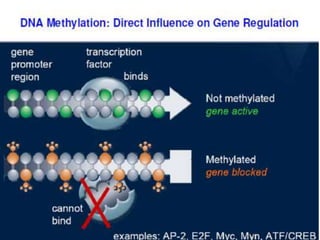
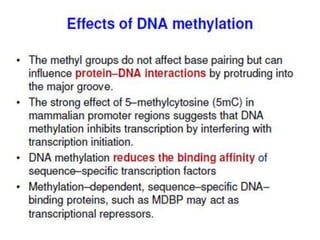
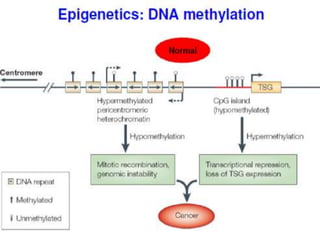
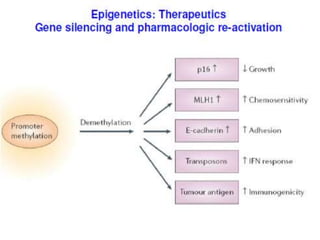
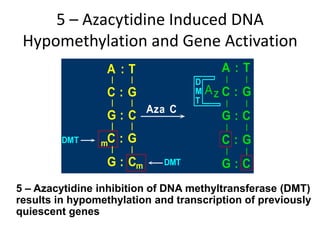
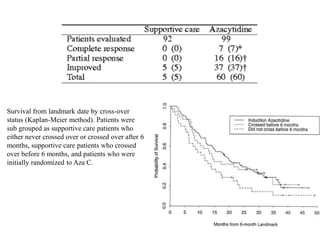
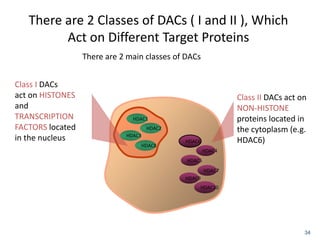
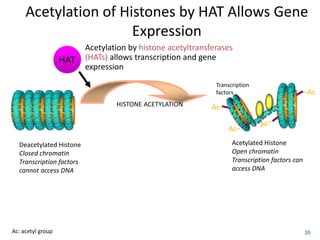
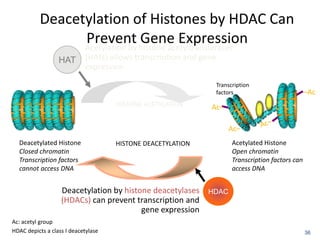
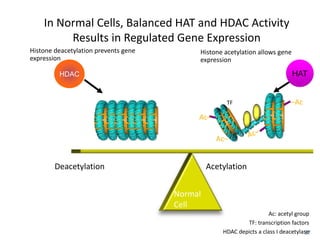
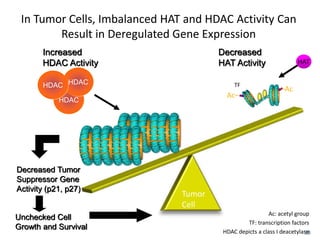
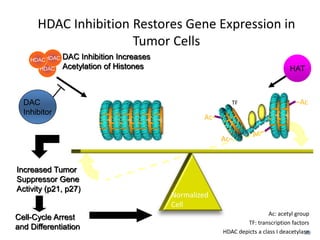
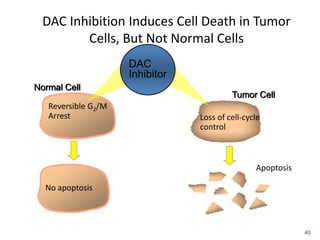
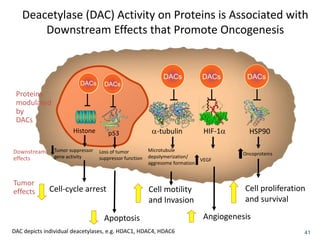
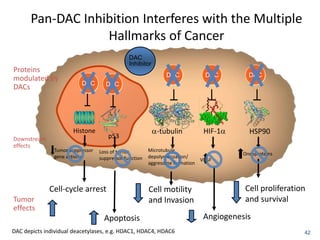
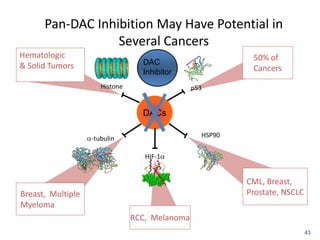
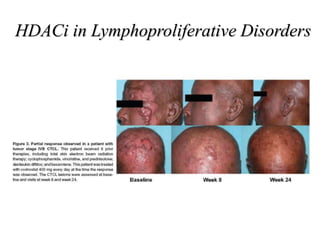
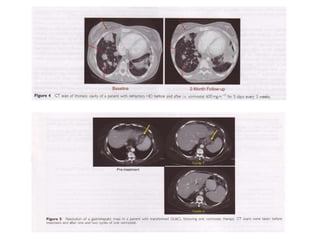
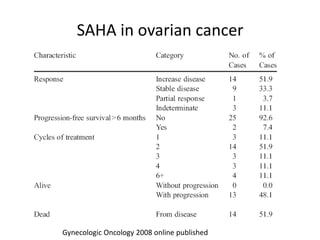
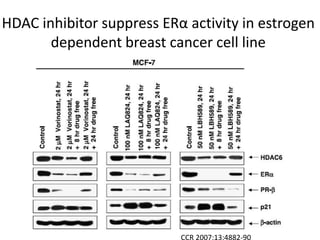
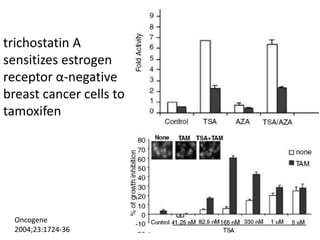
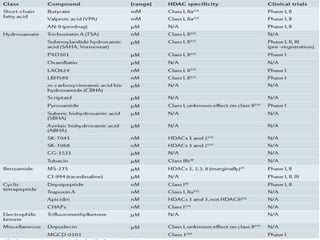
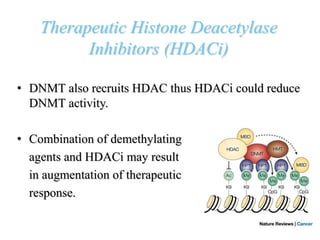
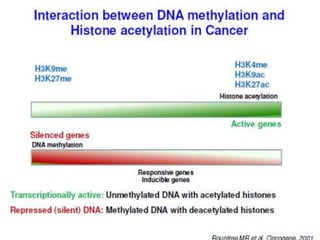
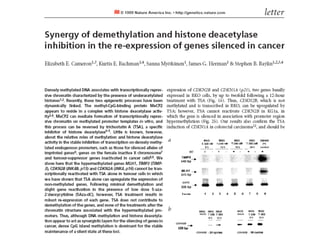
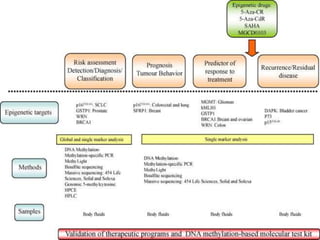
This document discusses epigenetics and cancer epigenetics. It defines epigenetics as heritable changes in gene expression that do not involve changes to DNA sequence. There are two forms of information in cells - genetic information encoded in the DNA sequence, and epigenetic information involving modifications like DNA methylation and histone acetylation. These epigenetic changes can contribute to oncogenesis along with genetic variations. The document discusses mechanisms of epigenetic changes like DNA methylation and histone acetylation/deacetylation and how they regulate gene expression. It provides examples of epigenetic changes in tumors and diseases and potential epigenetic therapies using demethylating and HDAC inhibitor drugs.