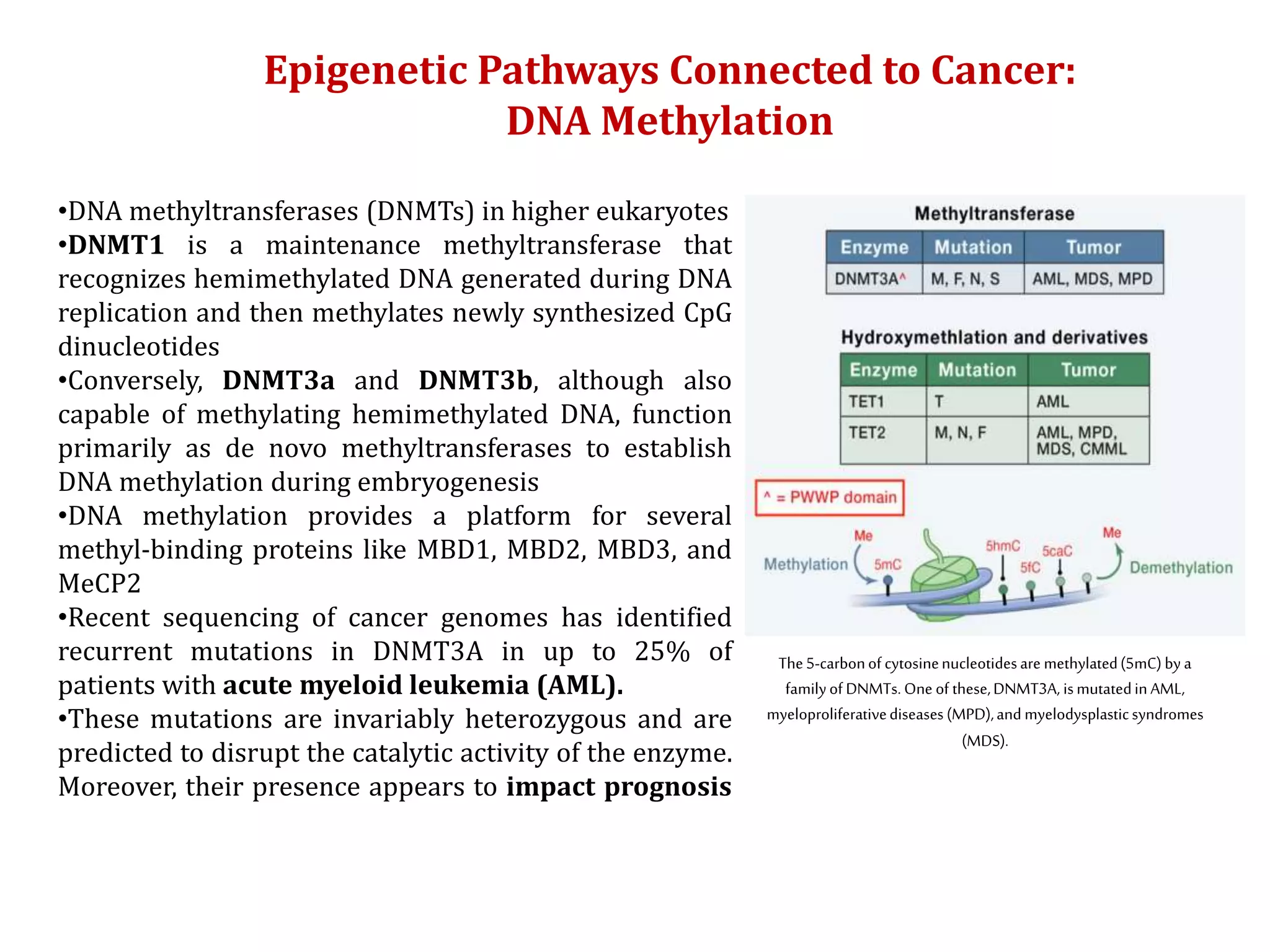
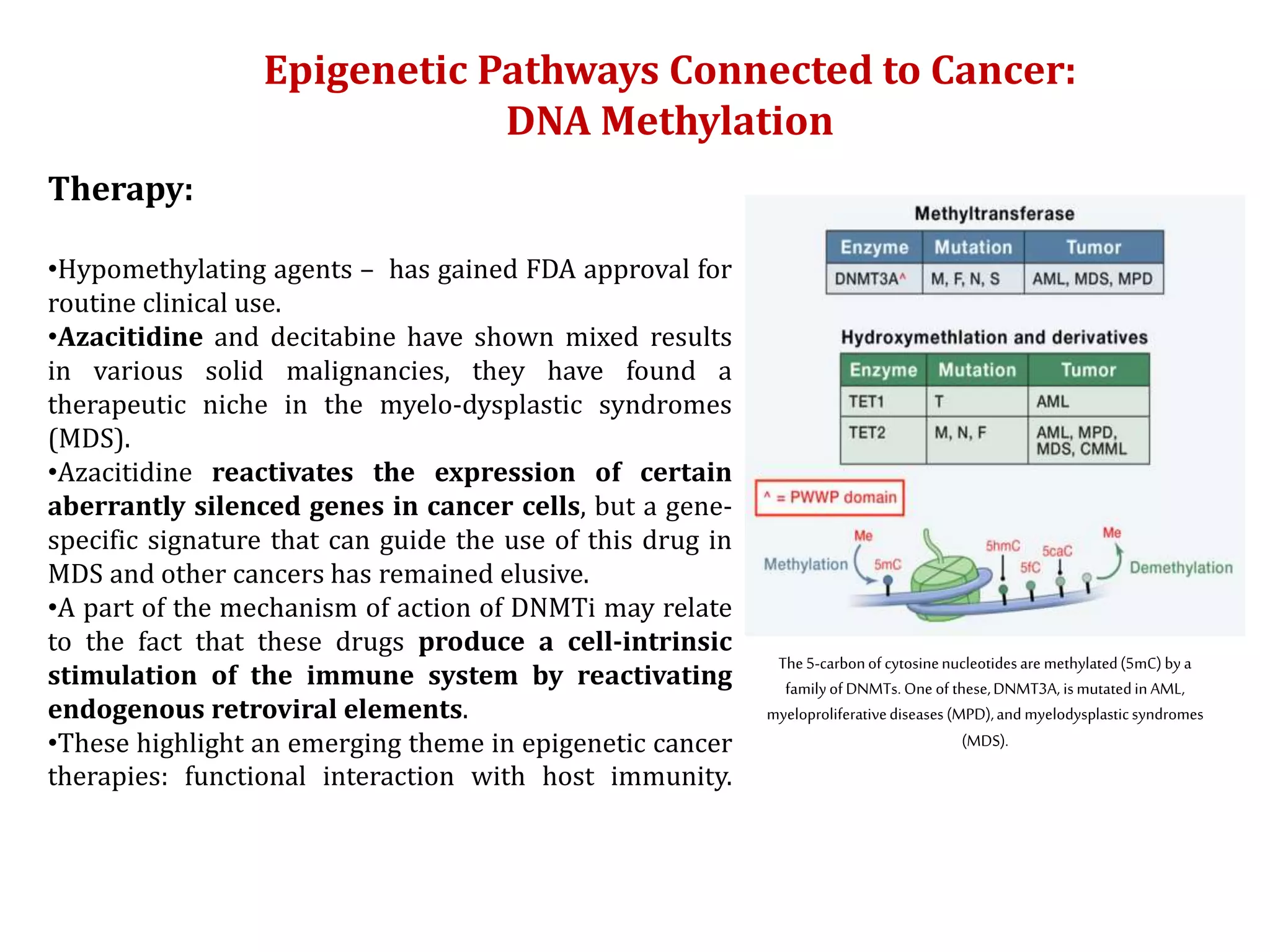
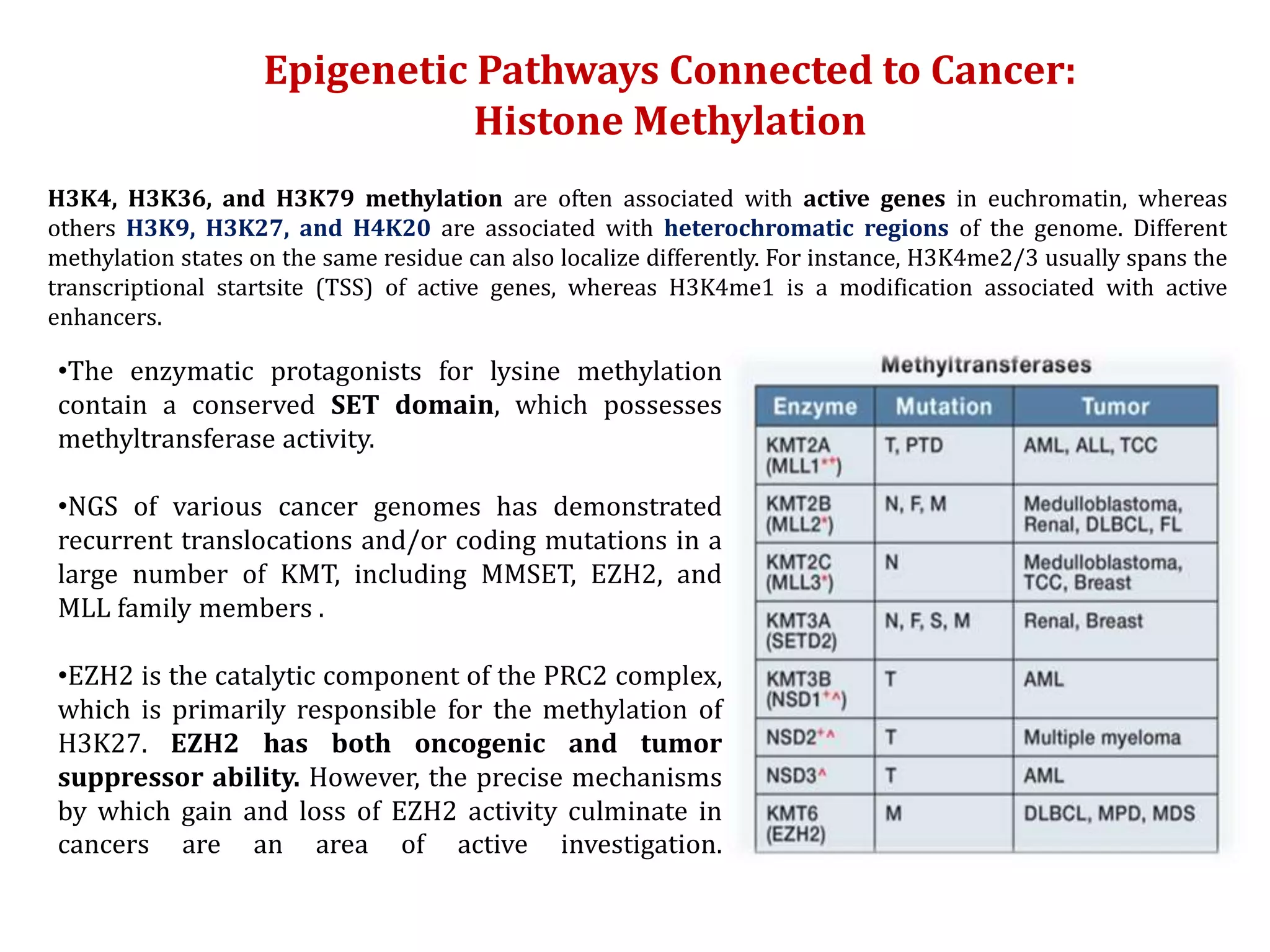
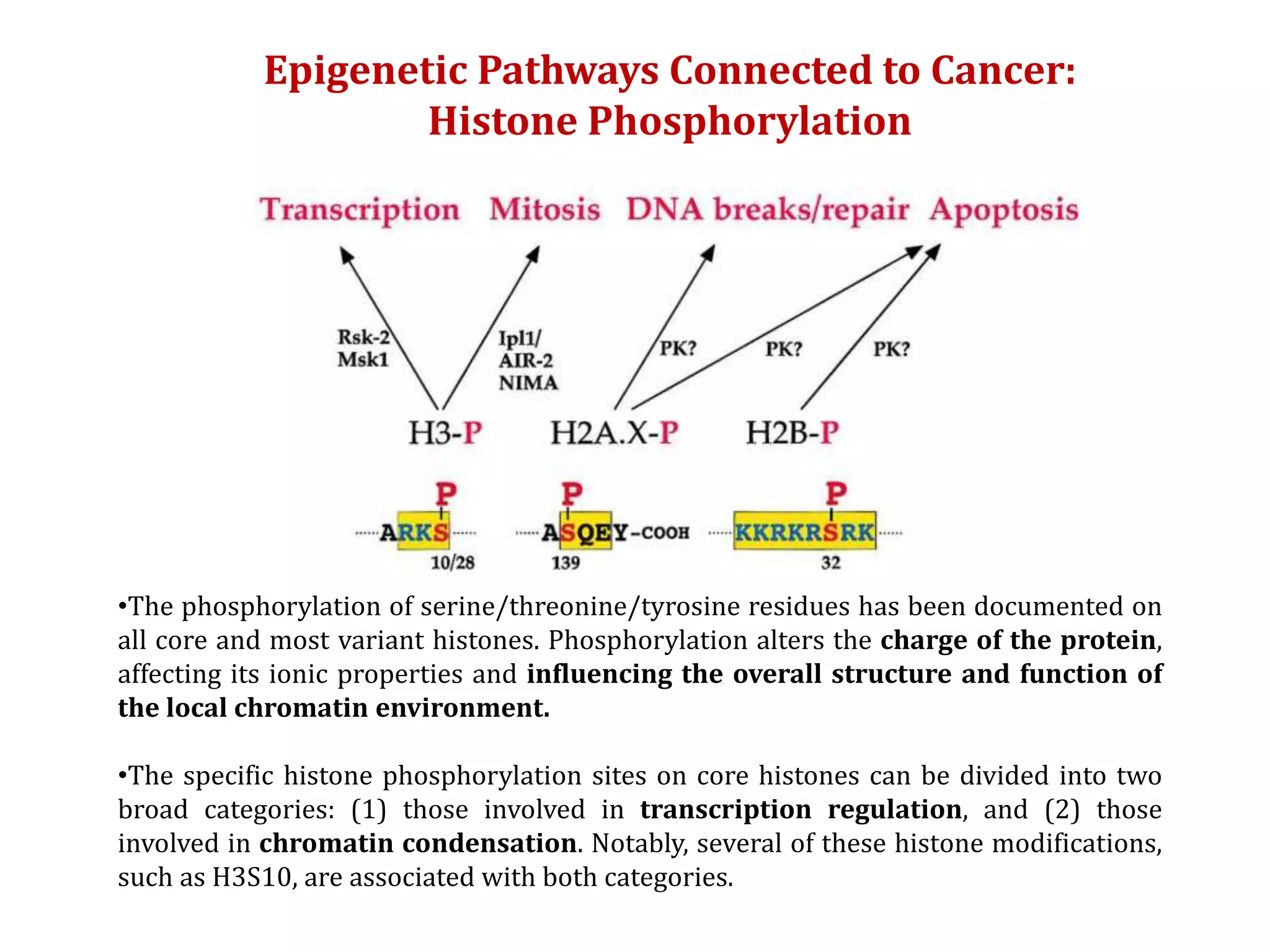
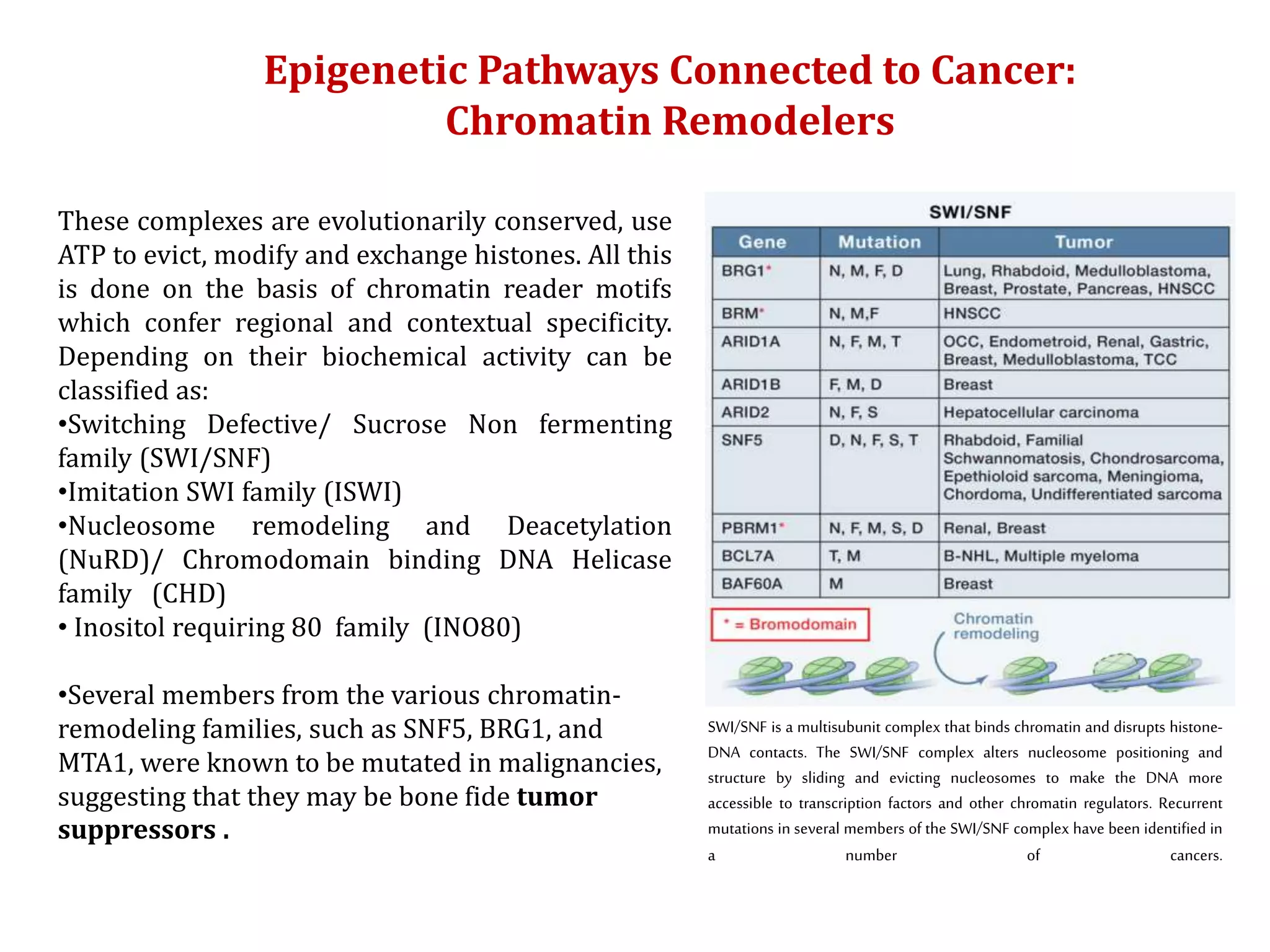
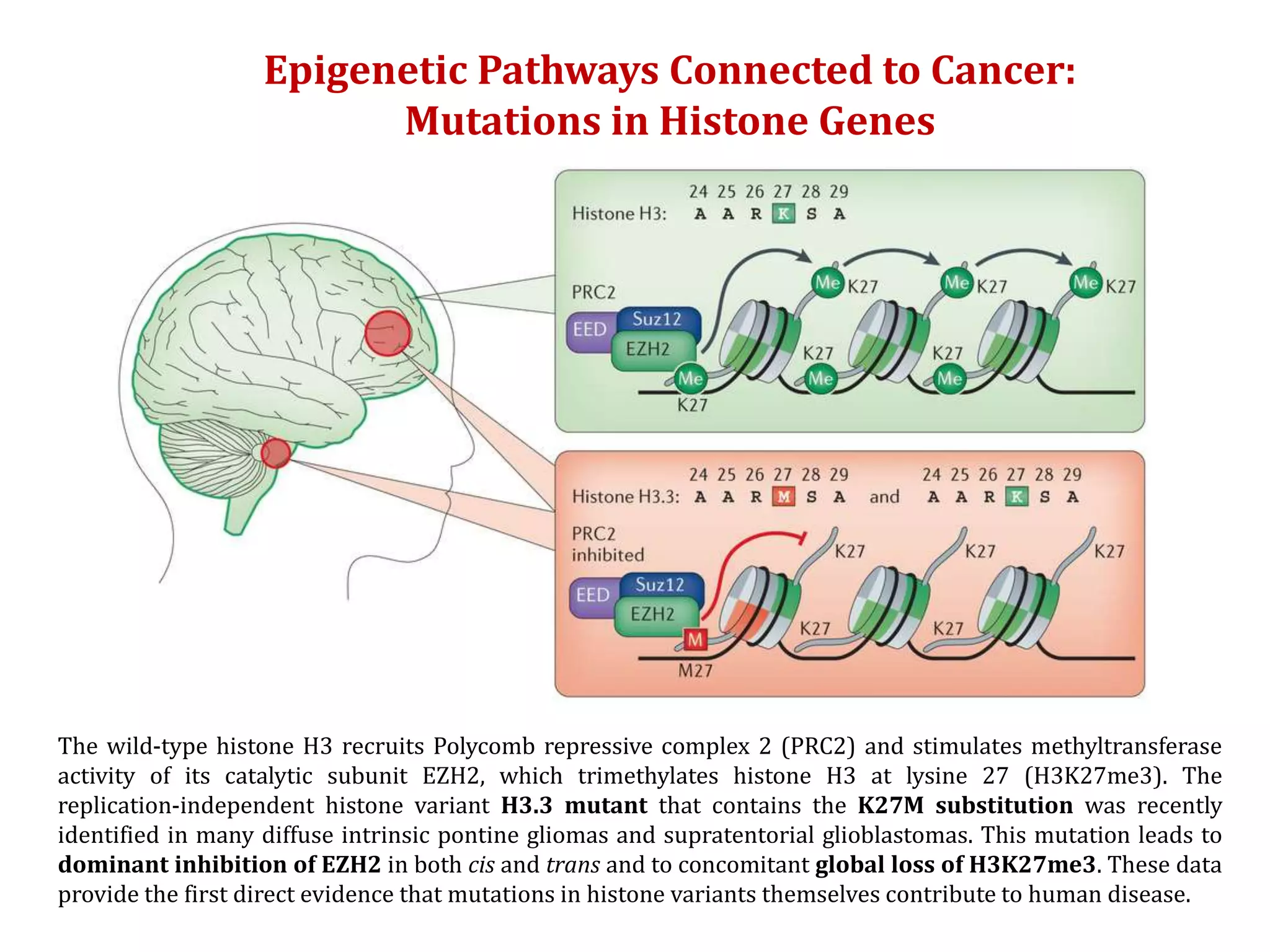
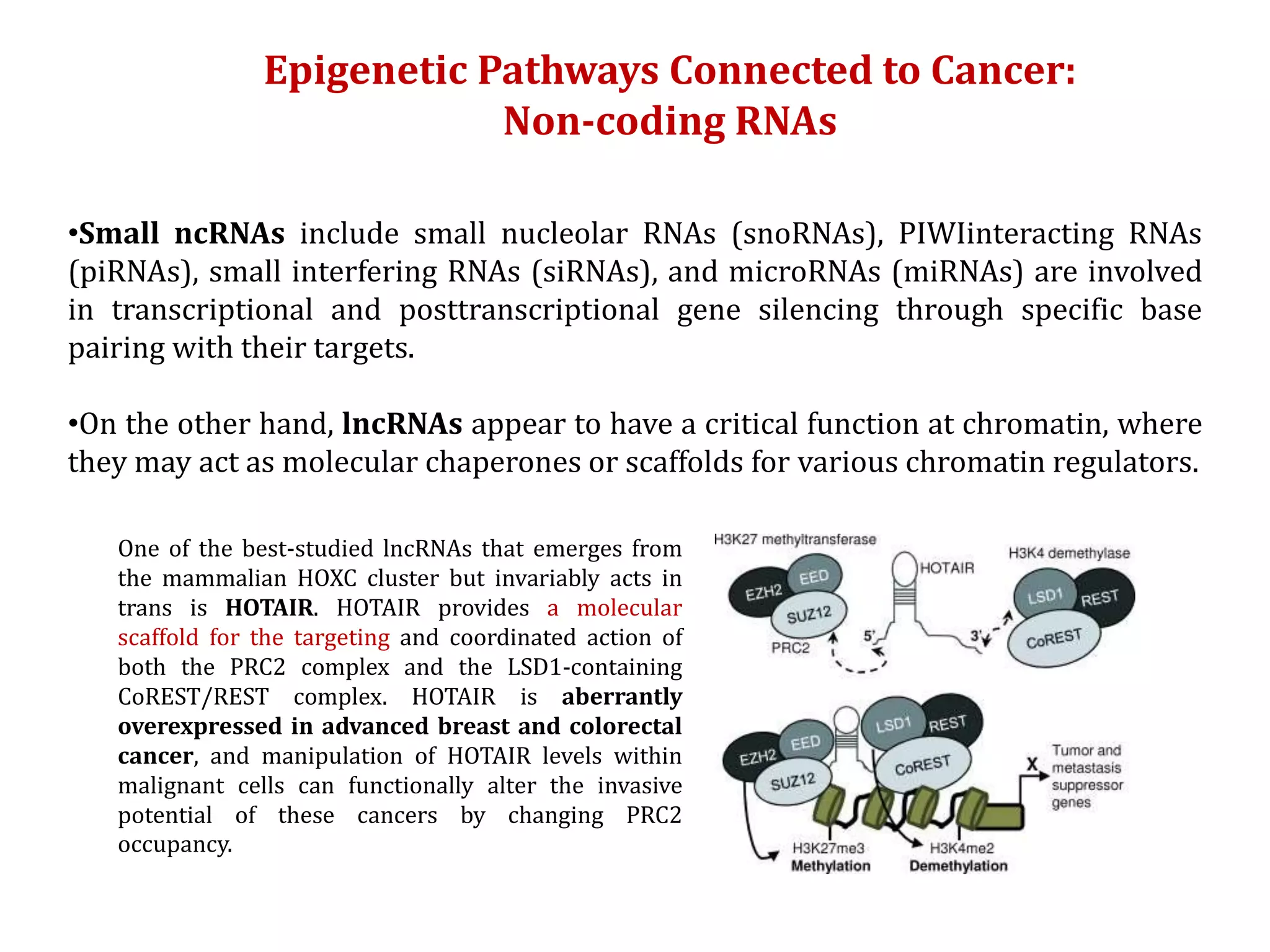
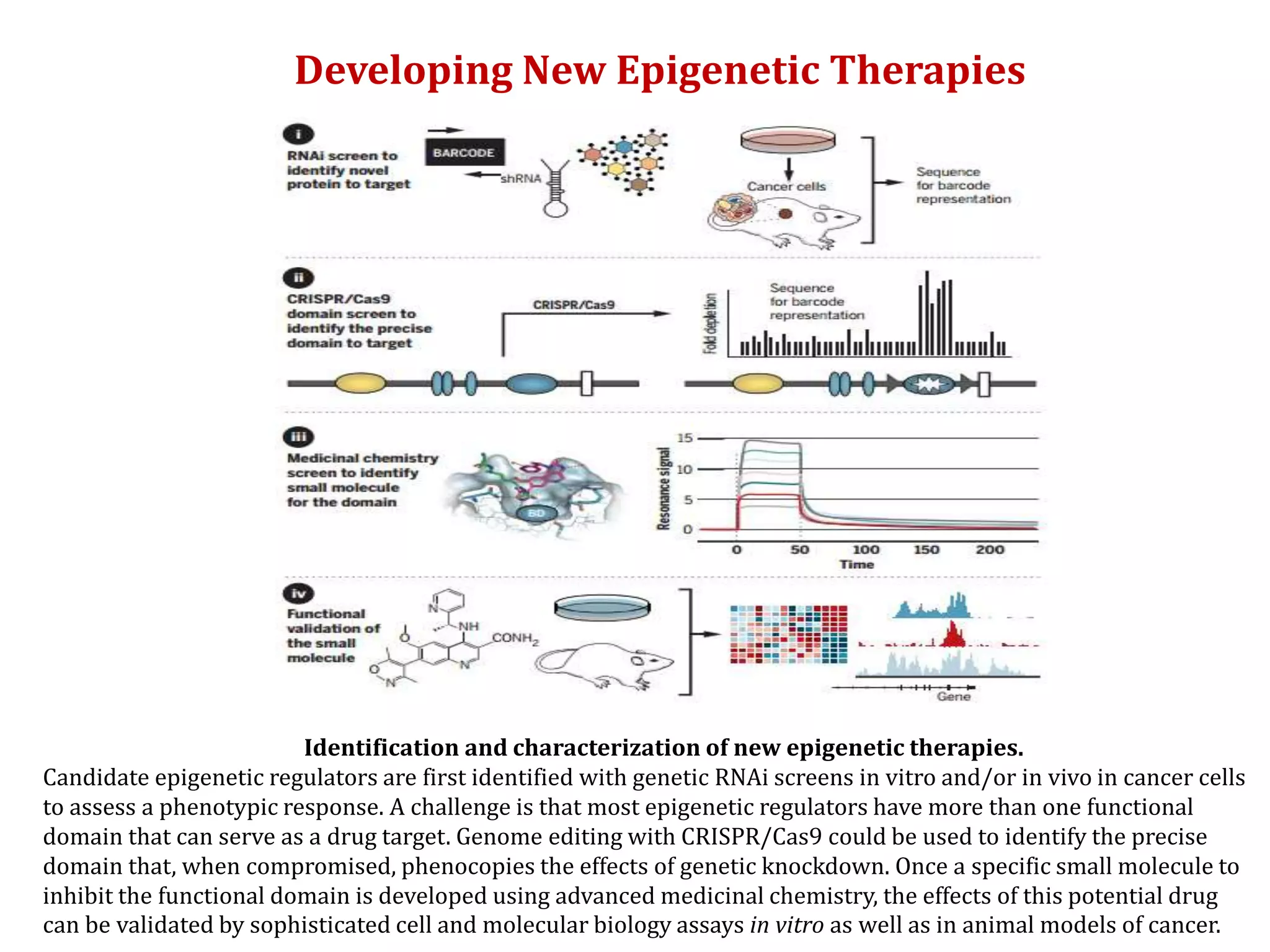
The document discusses cancer epigenetics, emphasizing heritable changes in cellular phenotypes not resulting from DNA sequence alterations. It details various epigenetic modifications, including DNA methylation and histone modifications, and their roles in cancer biology, noting the significance of mutations in specific genes like DNMT3A and TET2. The document also highlights therapeutic approaches involving hypomethylating agents and other targeted therapies based on epigenetic modifications.