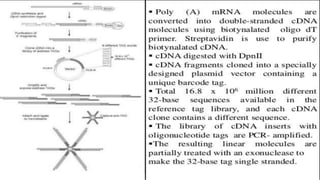
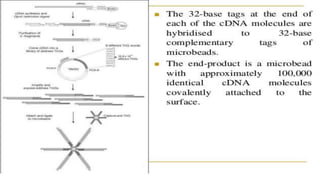
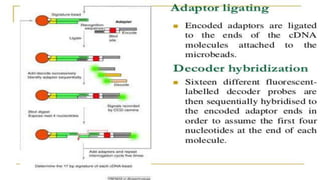
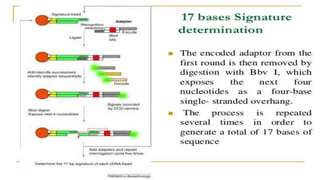
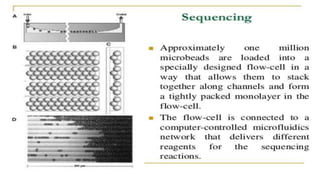
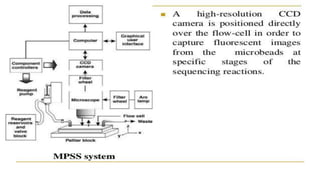
Lynx Therapeutics' Massively Parallel Signature Sequencing (MPSS) is an early high-throughput DNA sequencing technique. It works by attaching cDNA from an mRNA sample to beads, determining short sequence signatures from many beads in parallel, and using the signatures to count the number of individual mRNA molecules from each gene. This digital gene expression data allows MPSS to accurately quantify genes expressed at low levels by analyzing transcripts from virtually all genes simultaneously. The technique involves converting mRNA to cDNA, attaching oligonucleotide tags, PCR amplification on beads, and using fluorescent probes to determine short sequences in increments of four nucleotides from millions of beads in parallel.