1. Artificial neural networks (ANNs) are being used as a bioinformatics approach for gene prediction and genetic diversity analysis. ANNs consist of interconnected layers that learn from input to output.
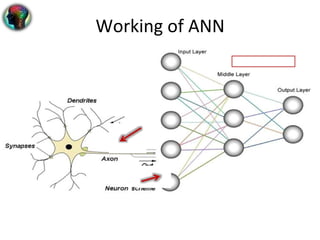
2. For gene prediction, a neural network is constructed with multiple input, hidden, and output layers. The input is a gene sequence and output is exon probability. Weights between layers are adjusted during training to recognize patterns.
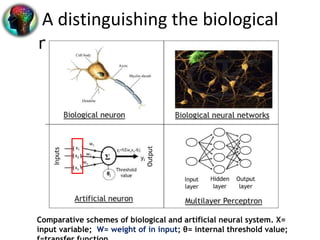
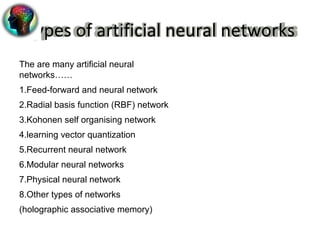
3. ANNs have advantages over traditional statistical methods as they can model more complex data relationships without requiring detailed system information. Different ANN types exist for various applications in bioinformatics.