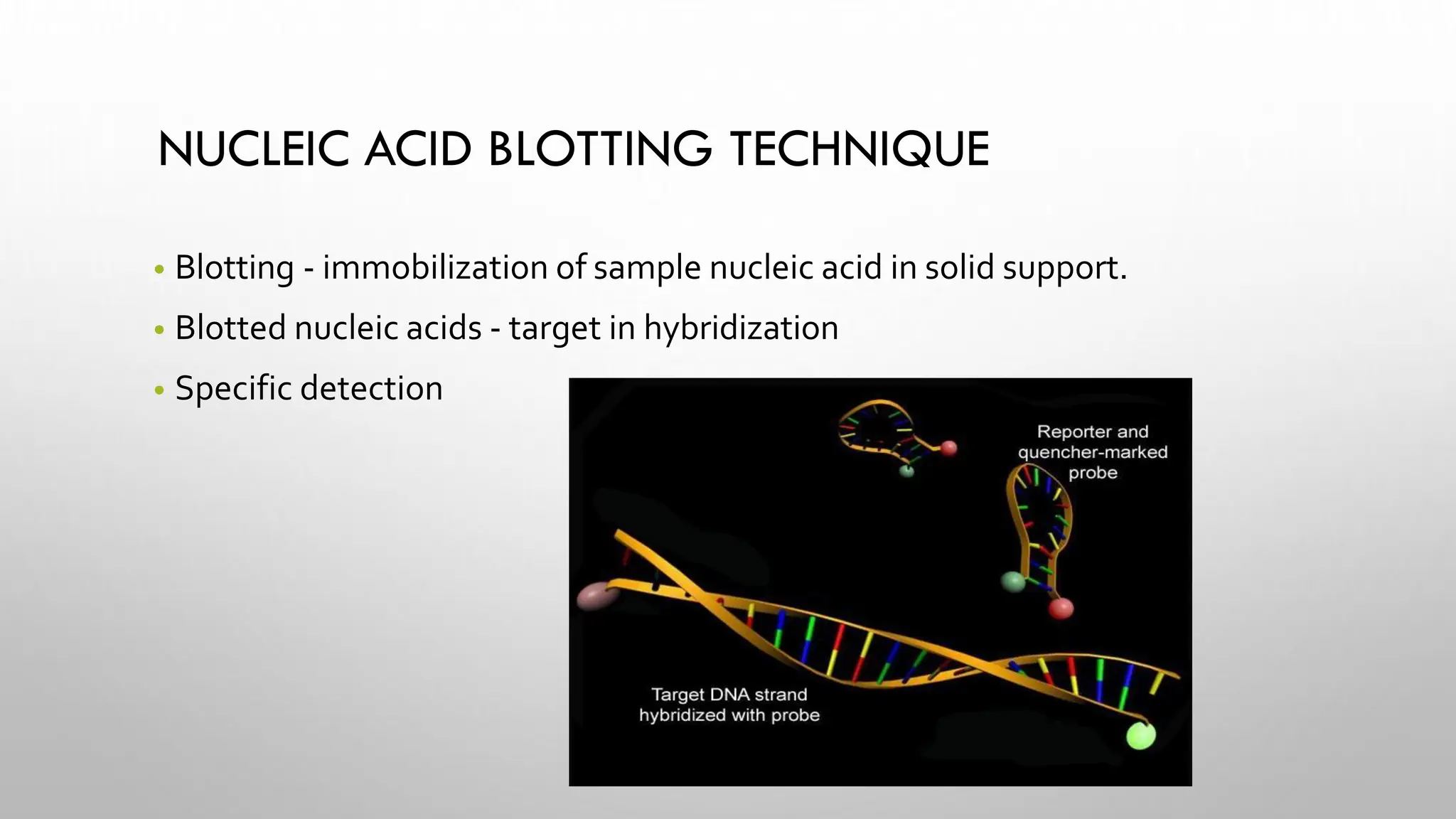
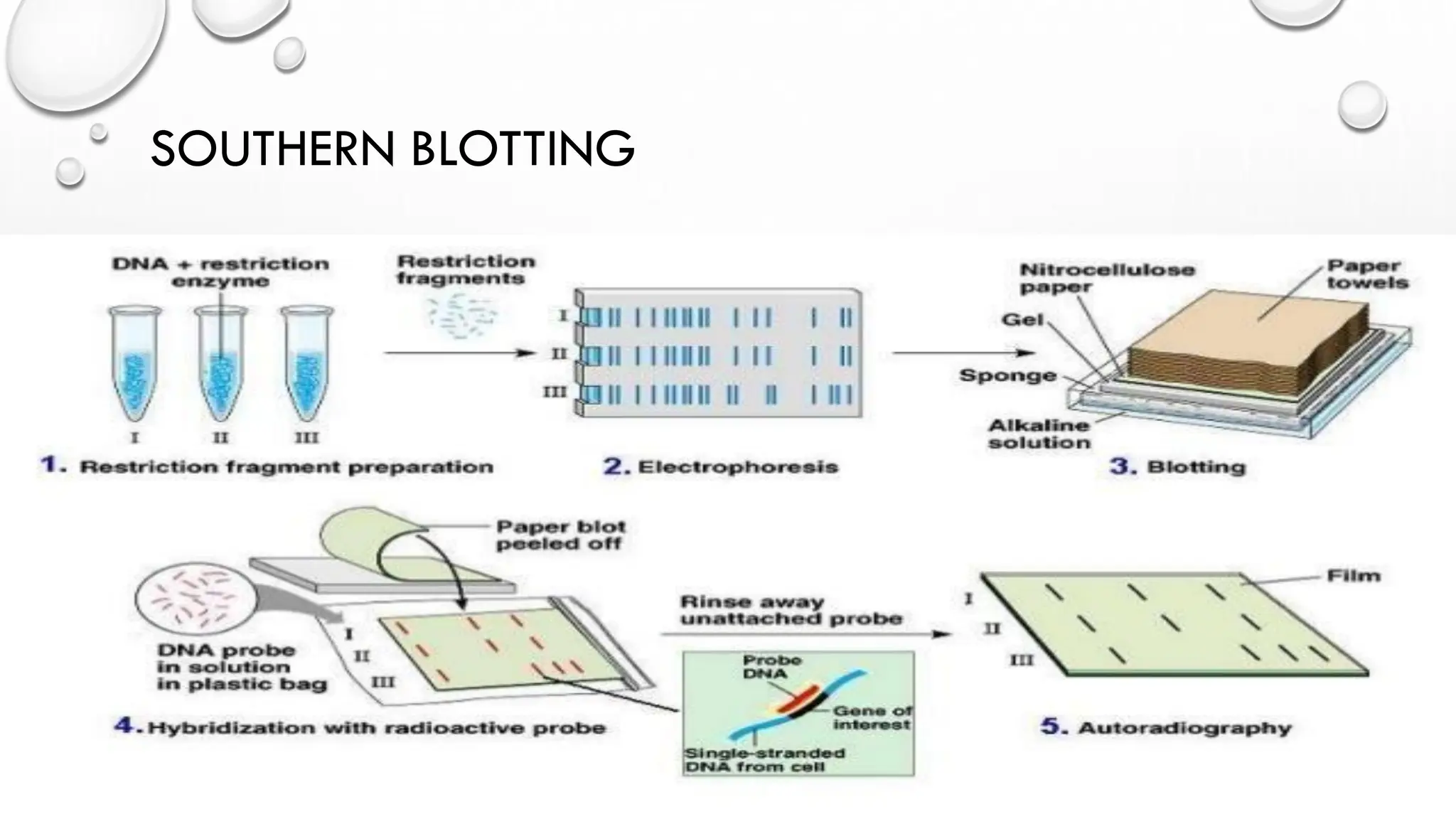
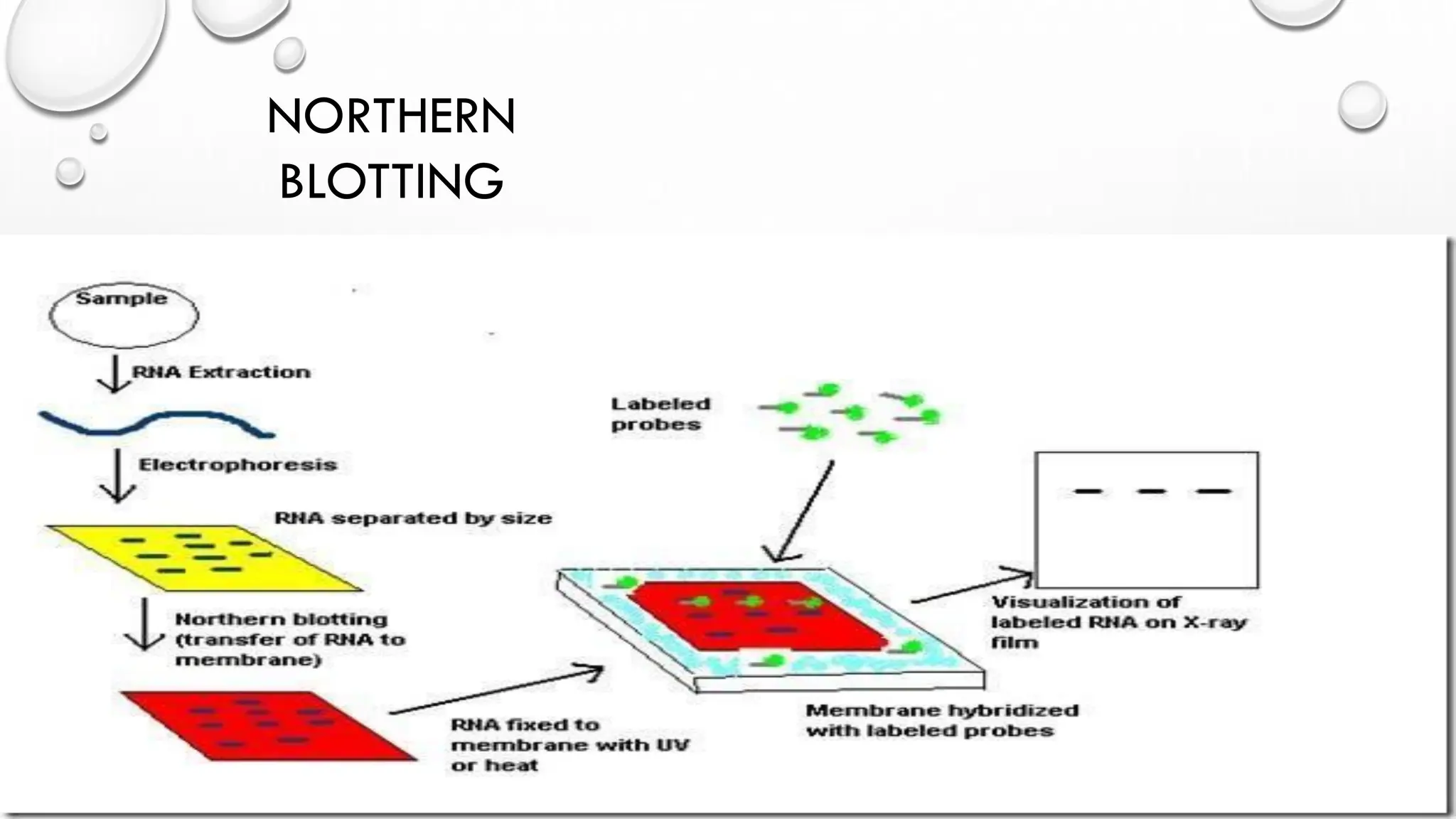
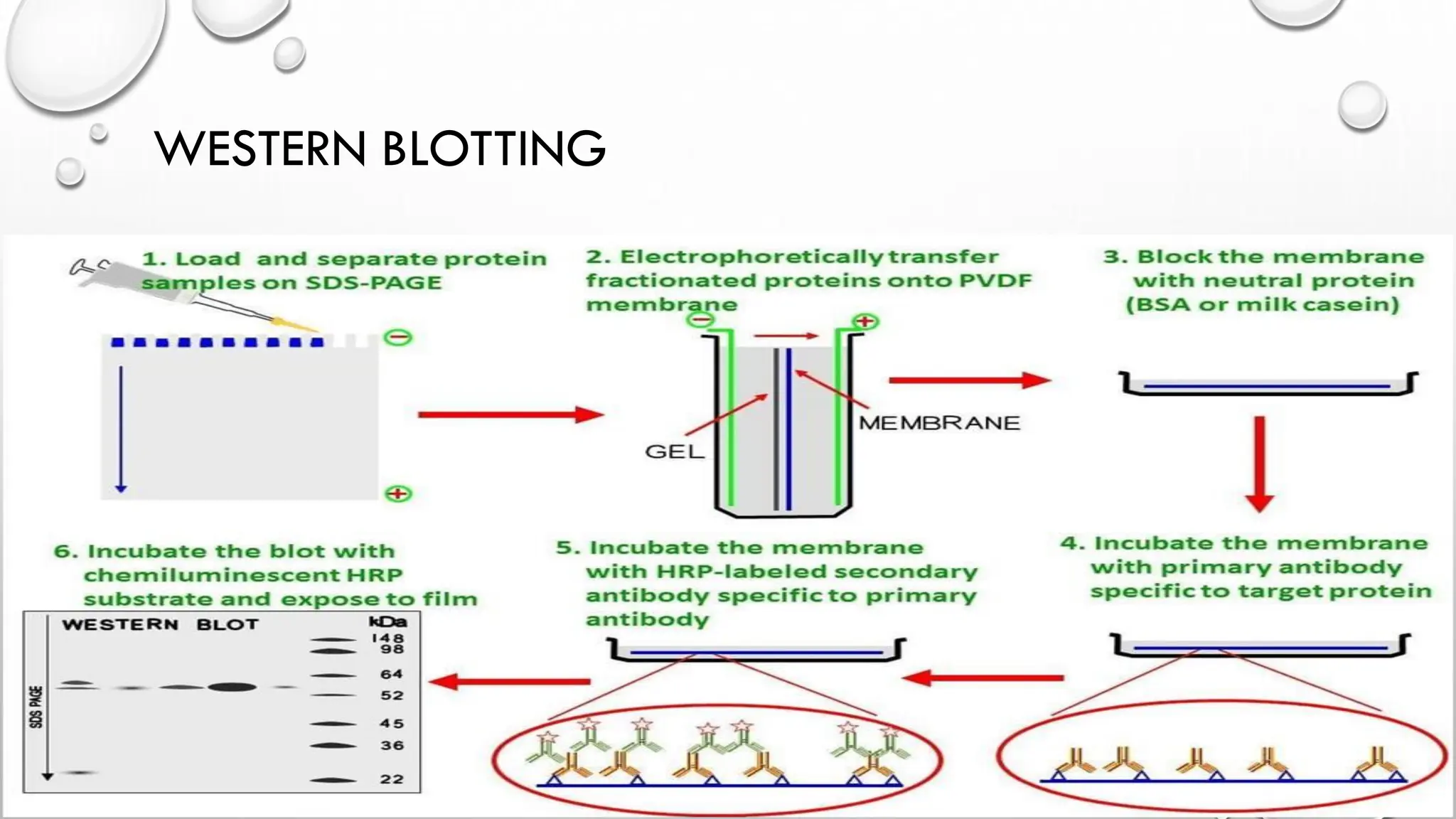
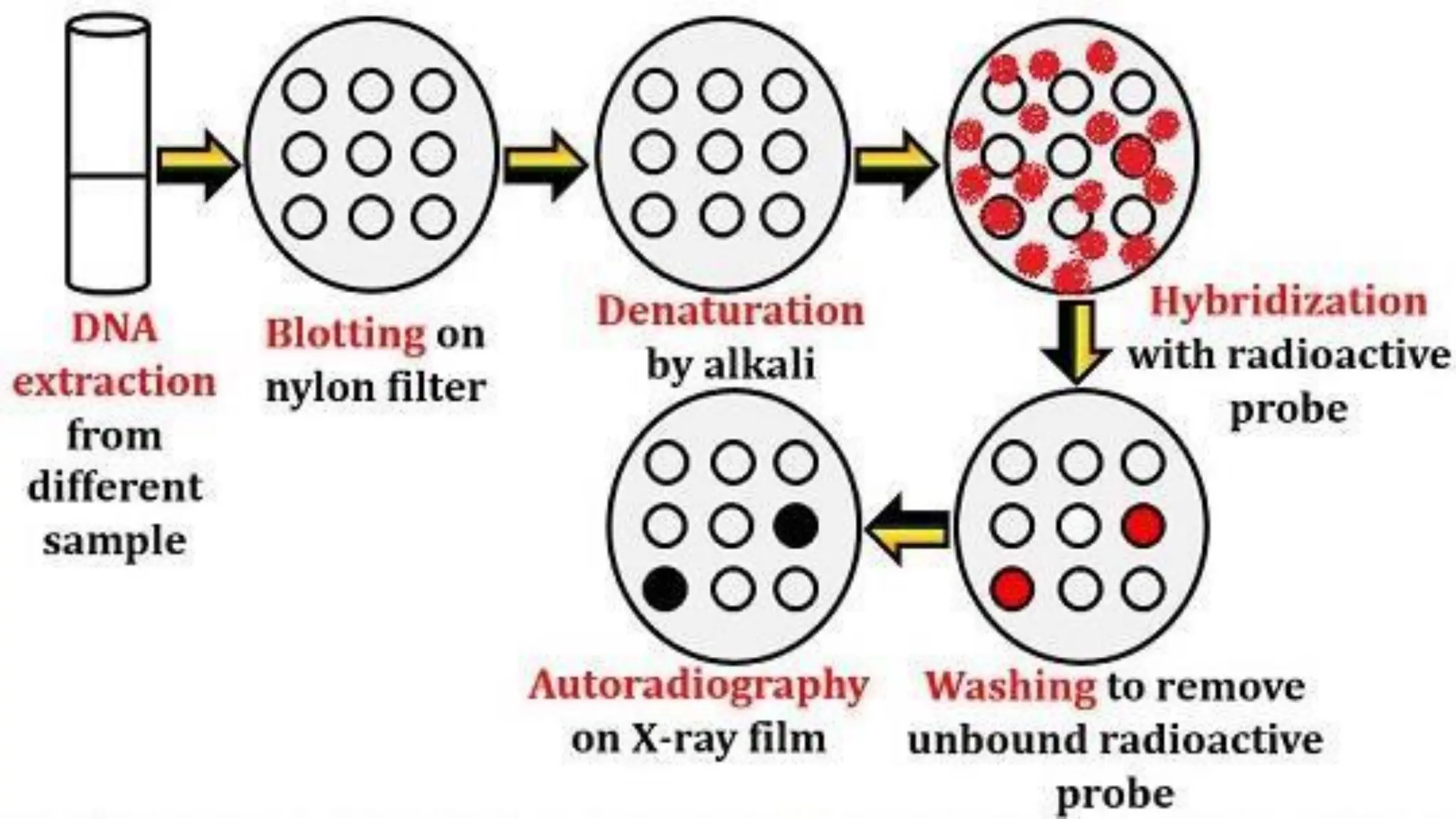
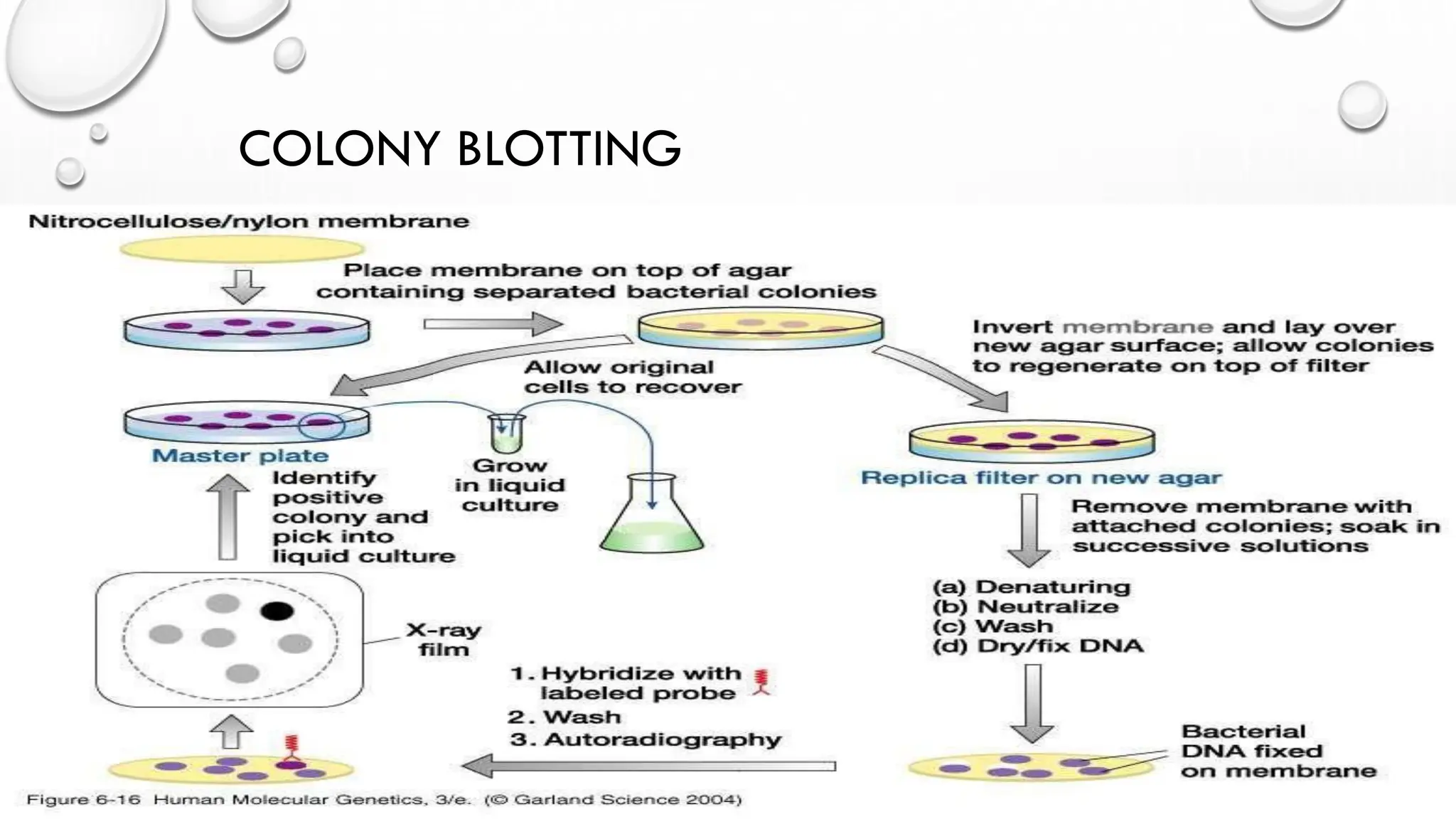
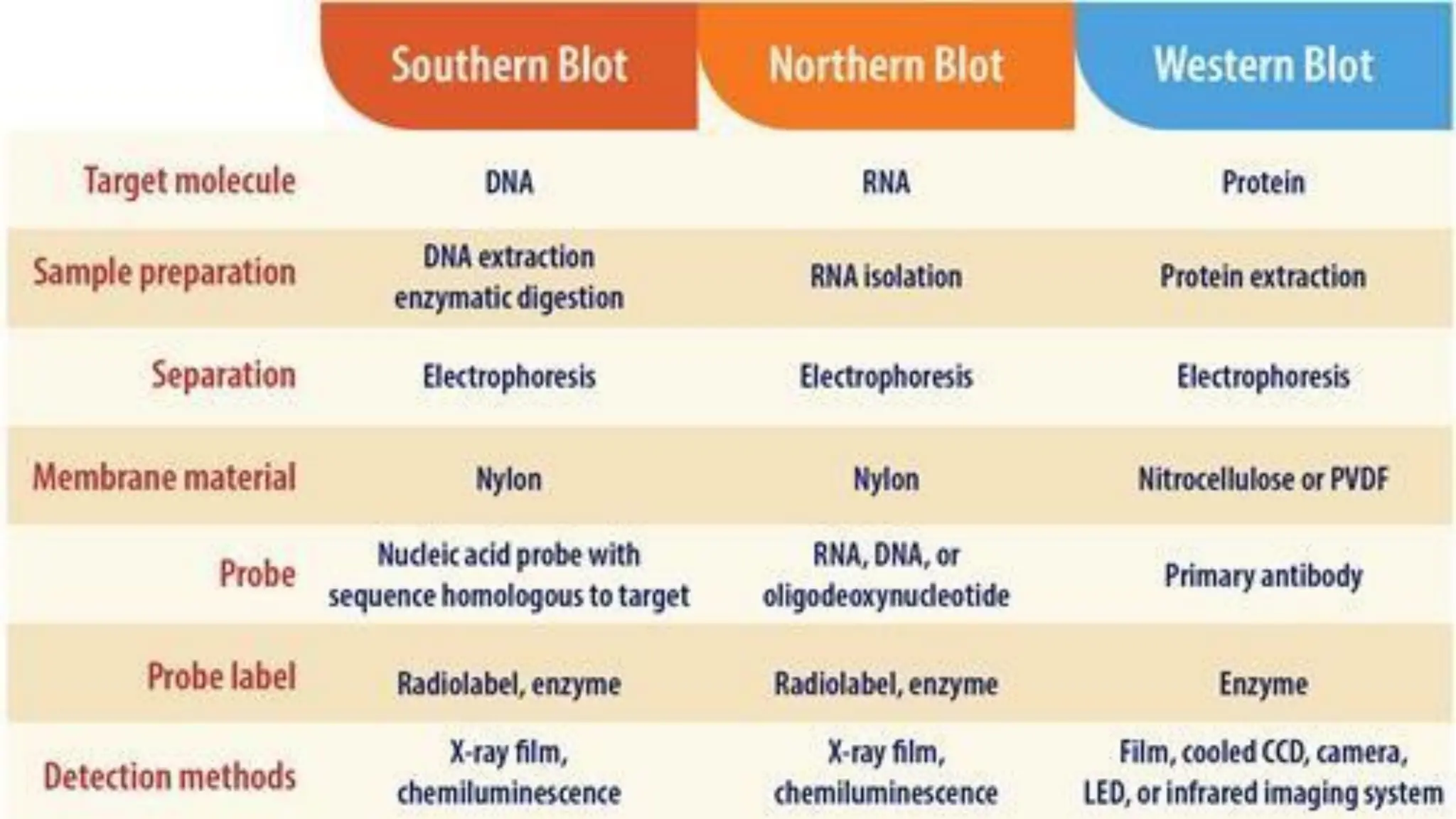
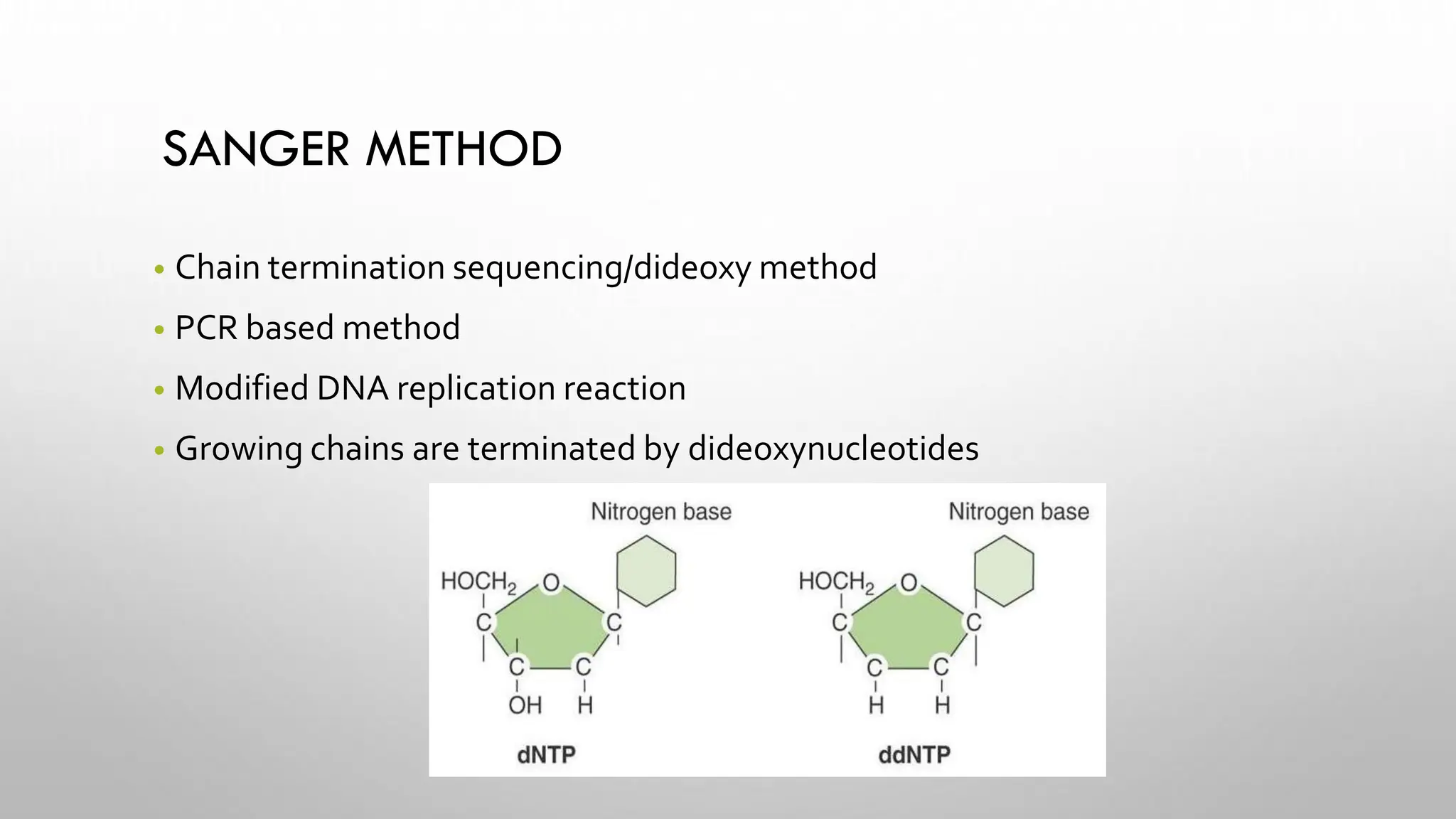
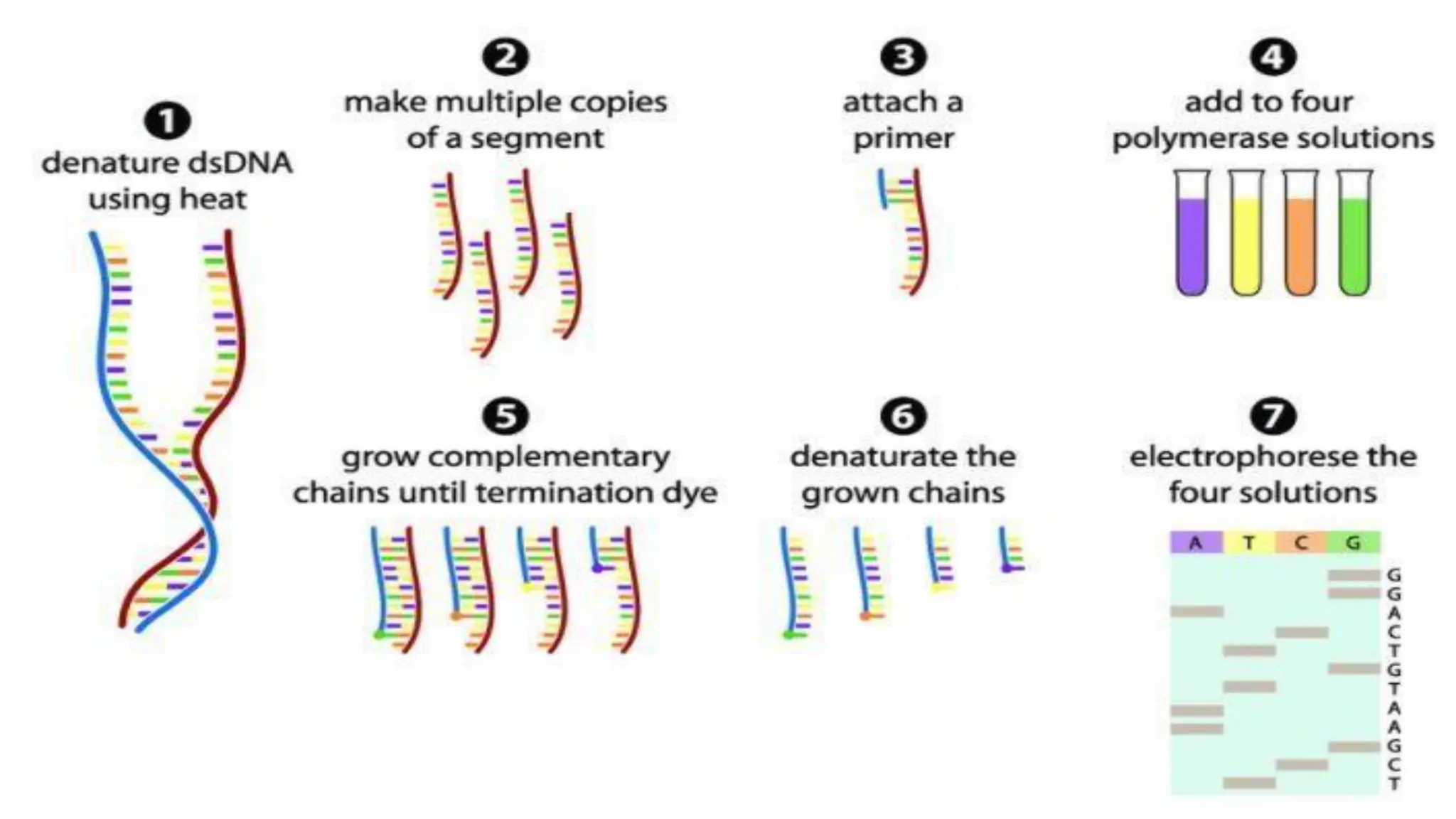
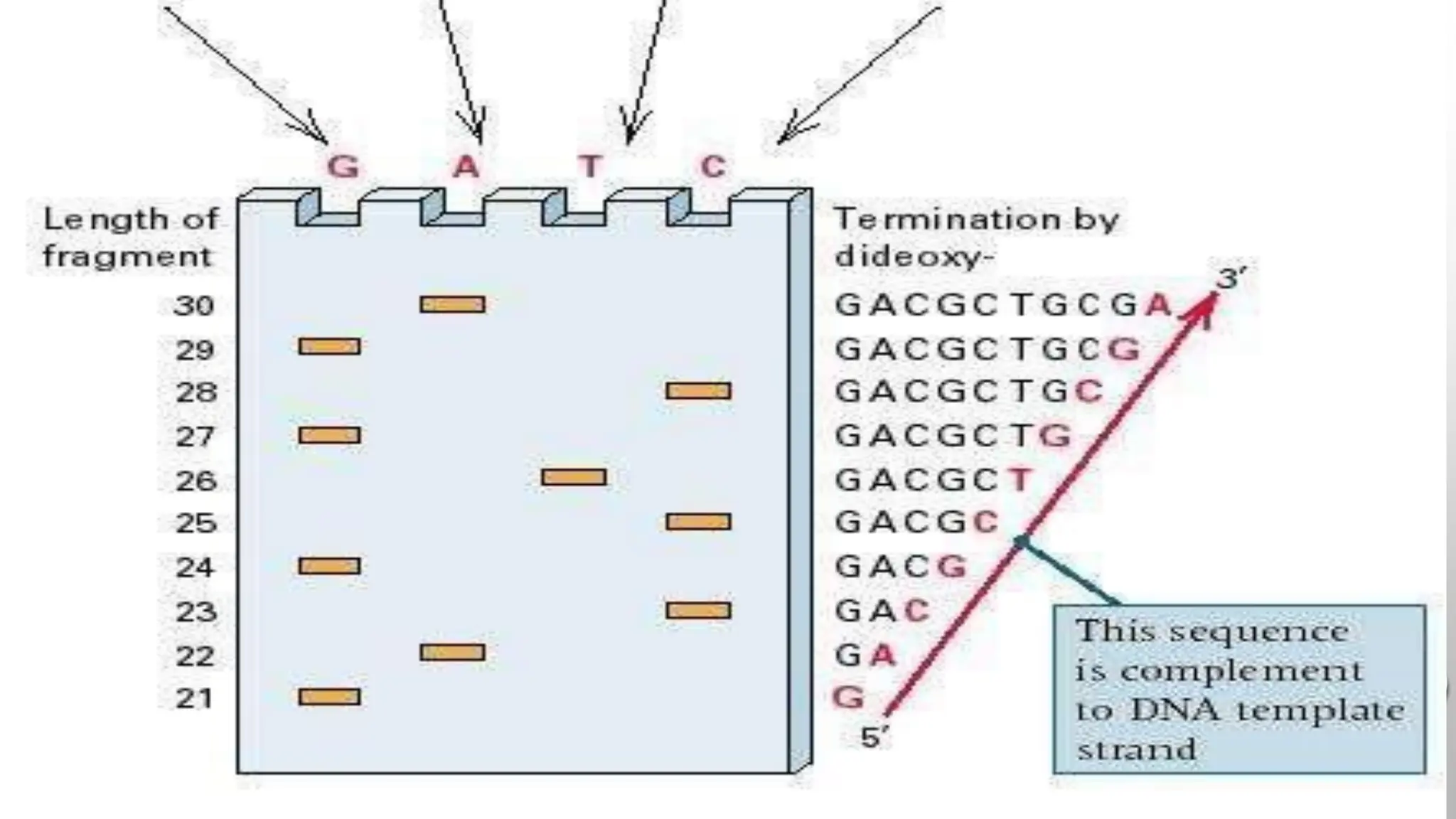
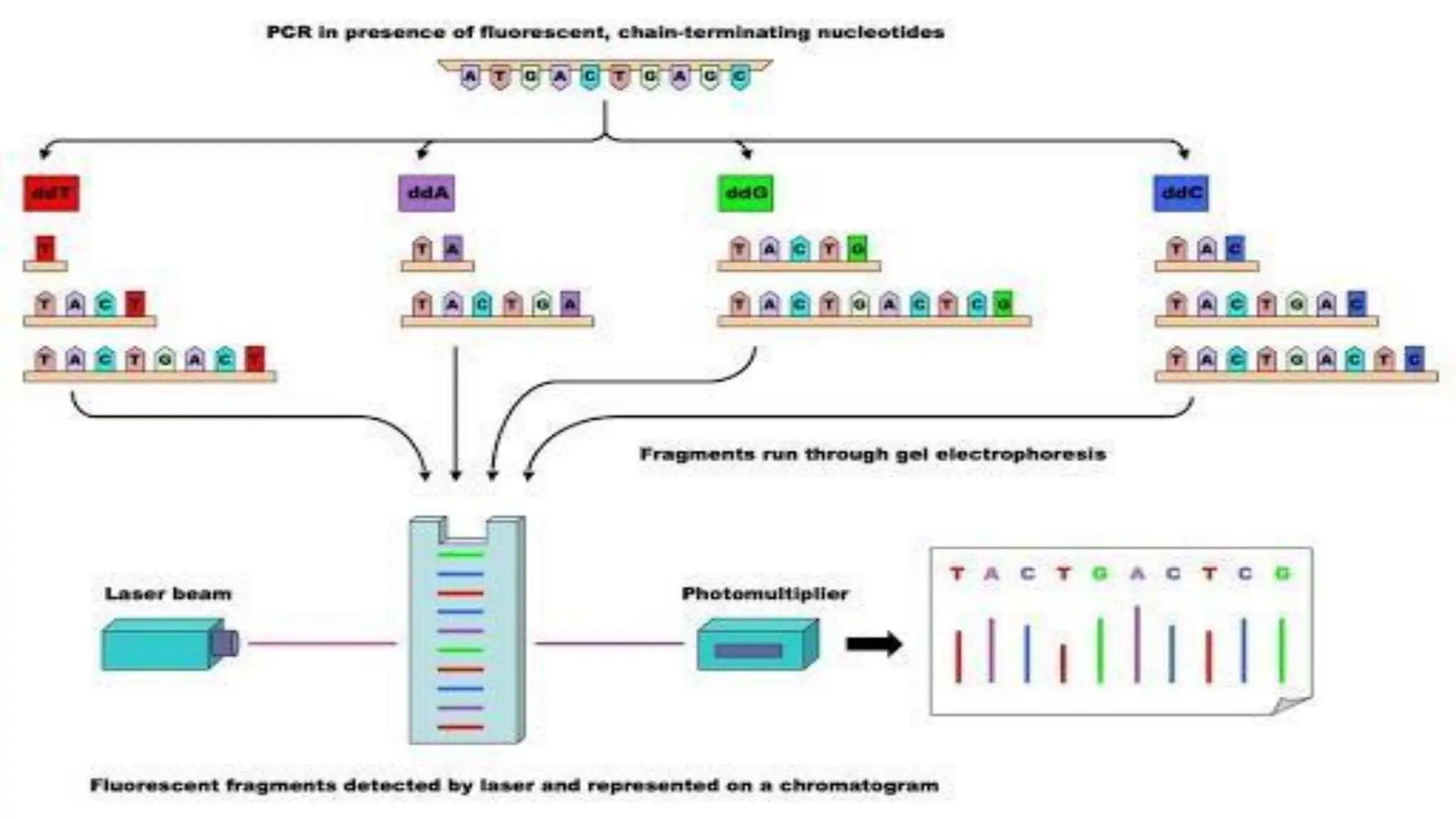
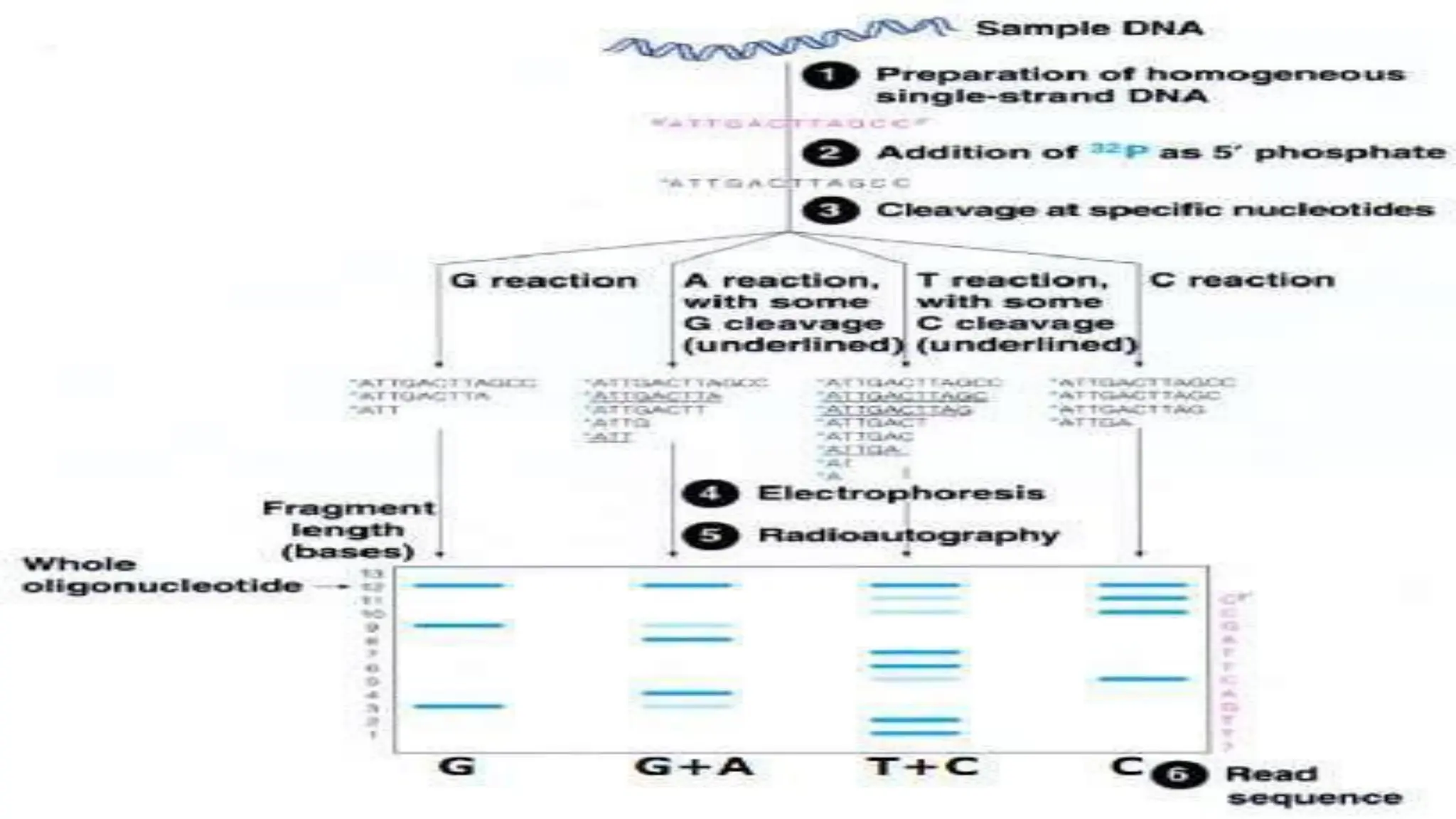
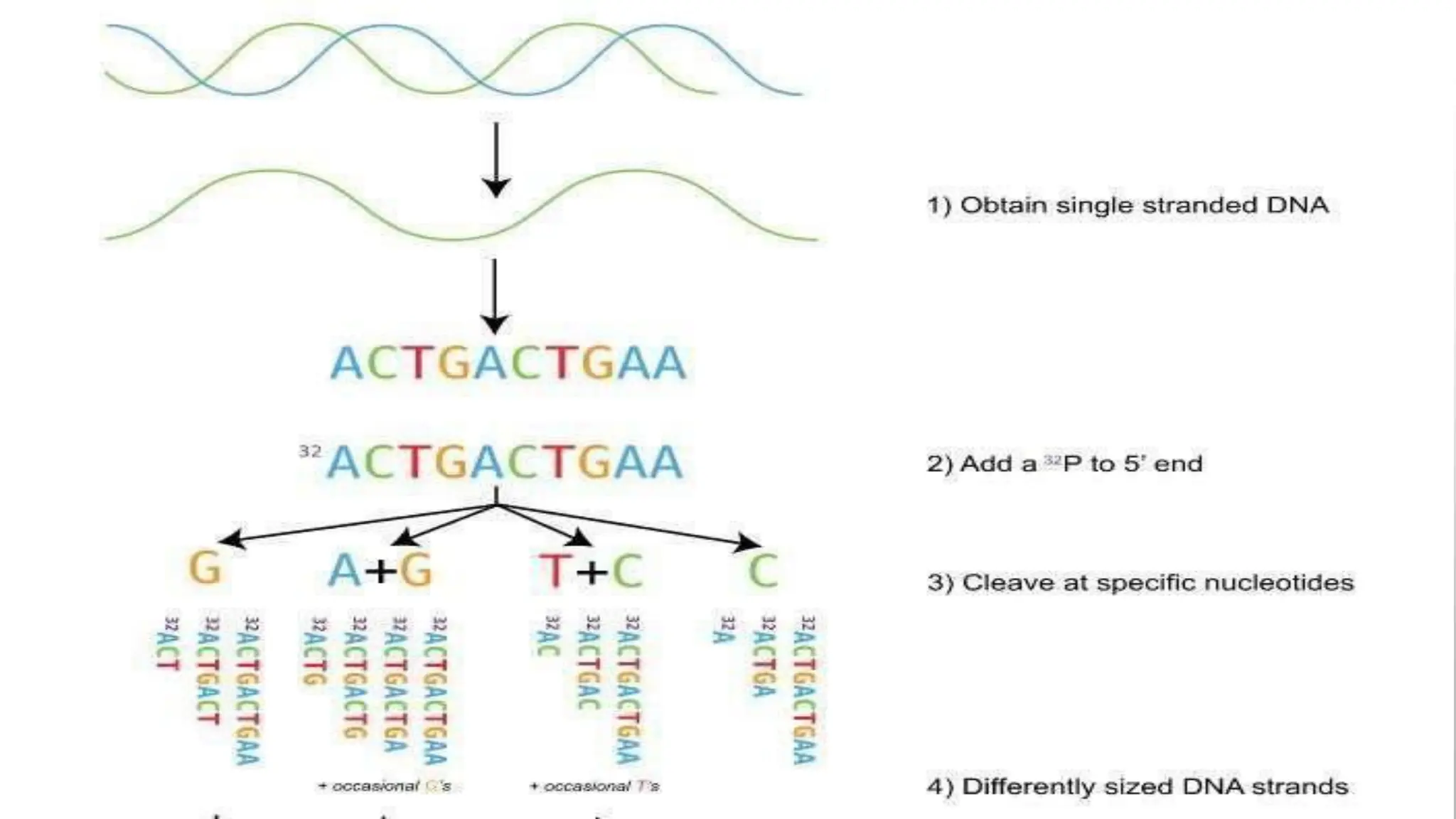
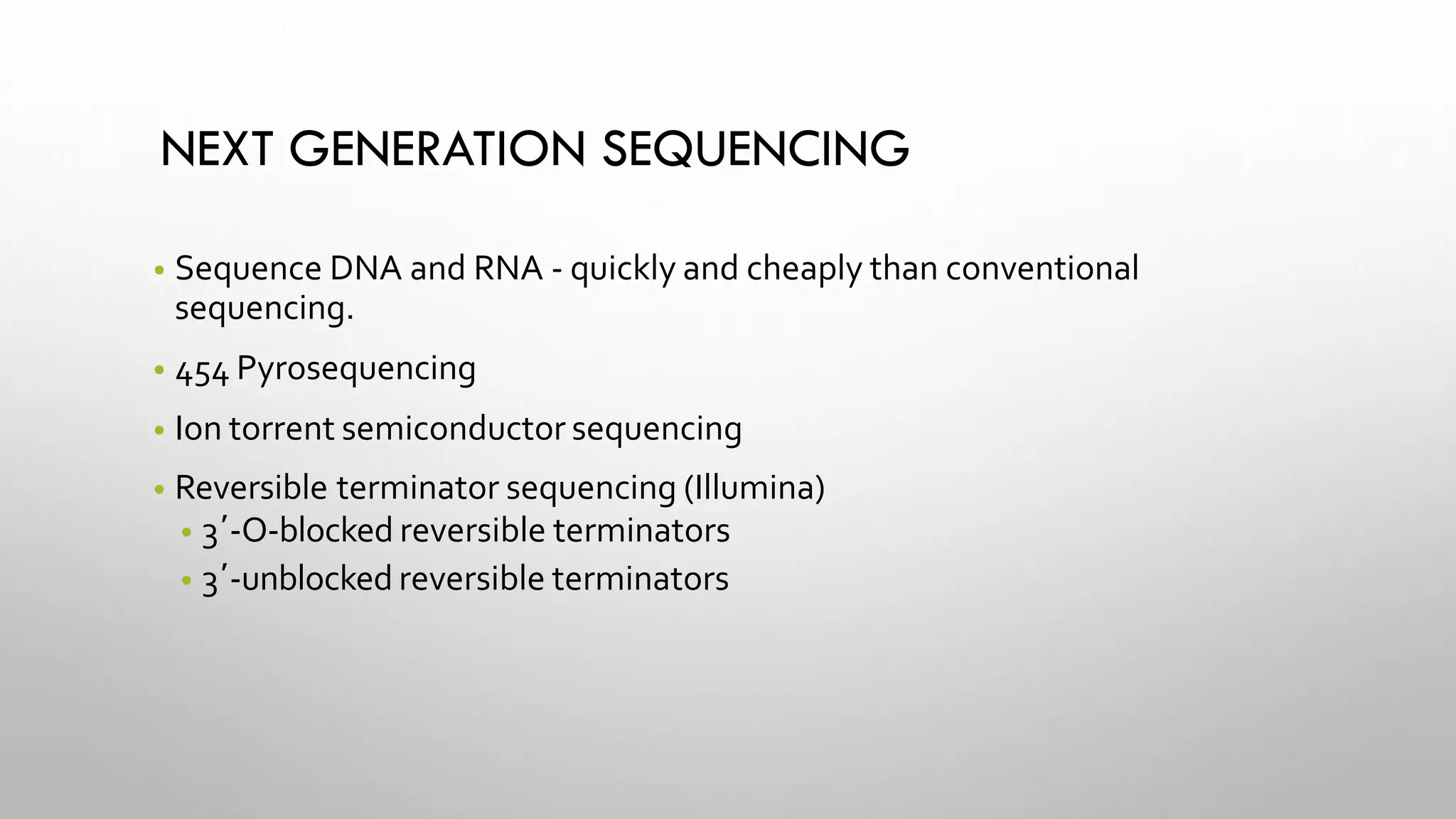
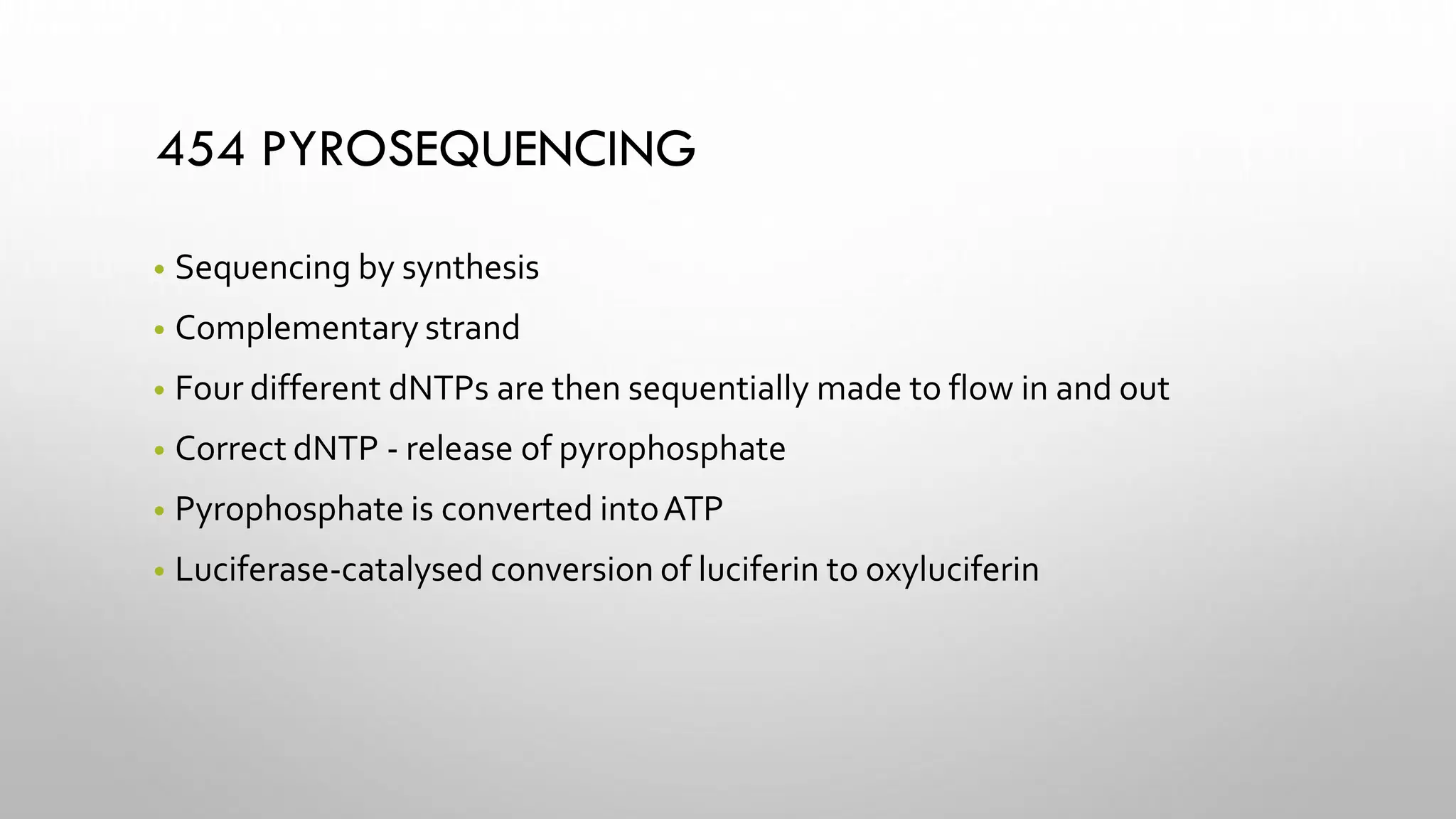
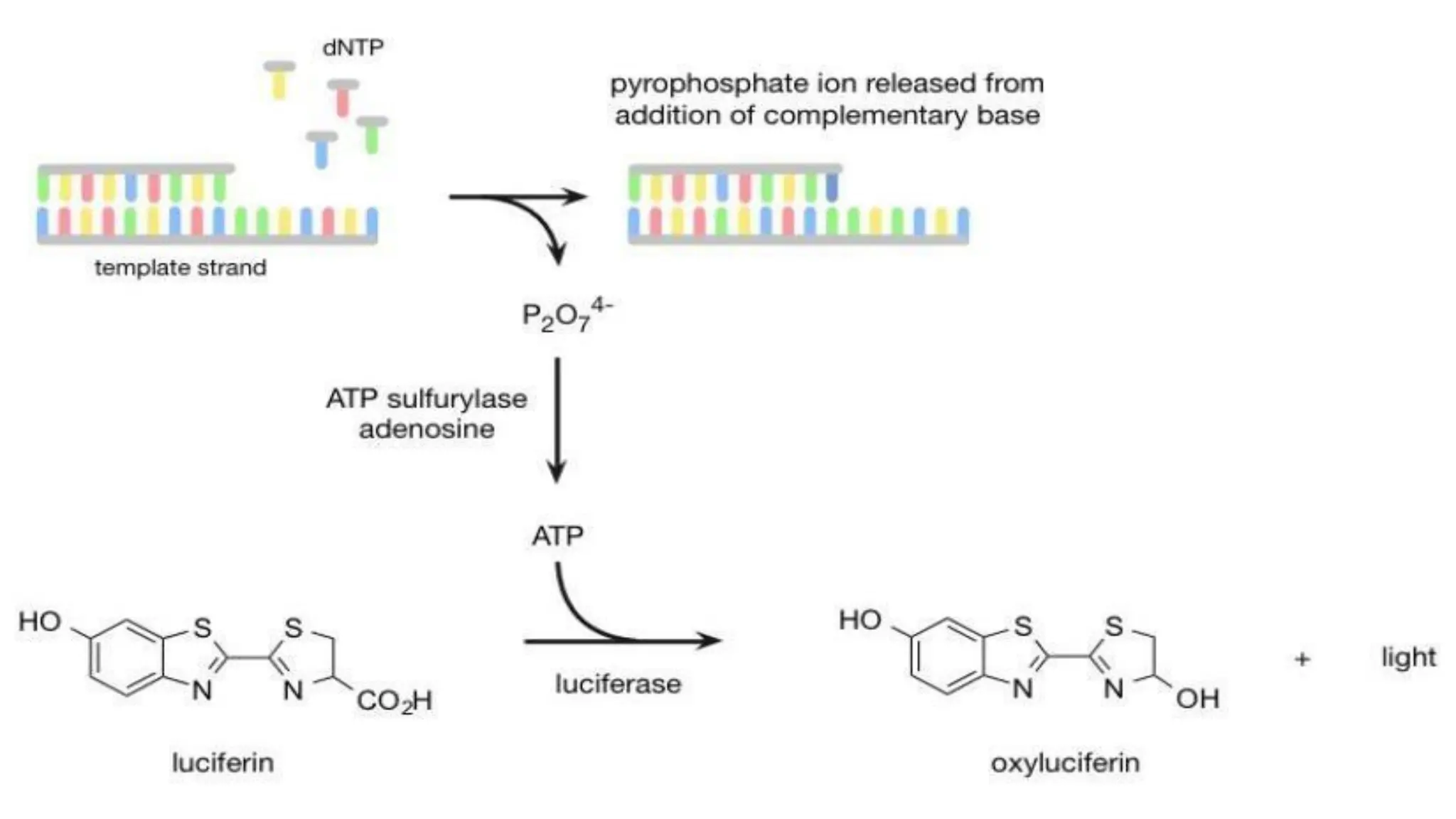
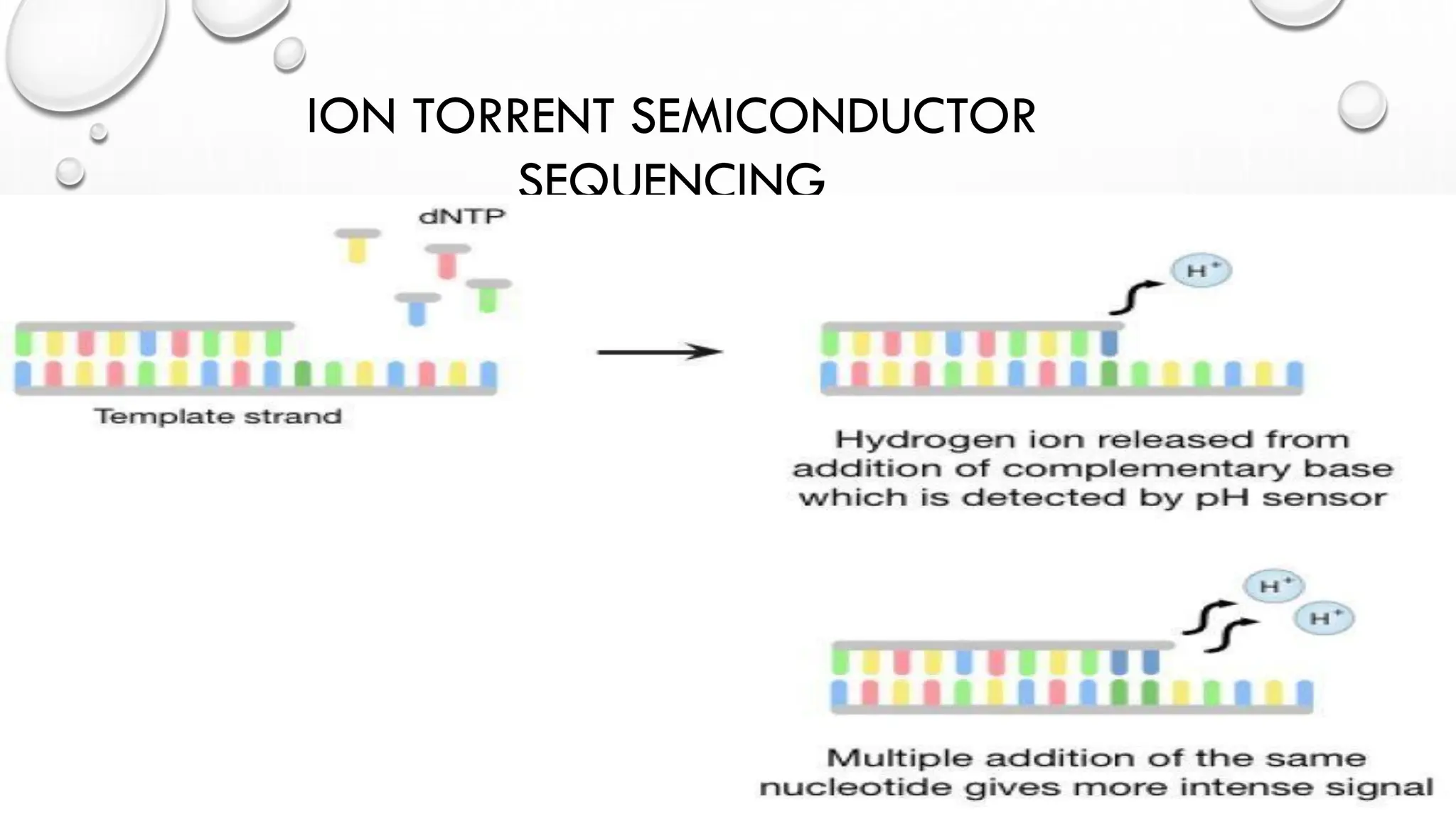
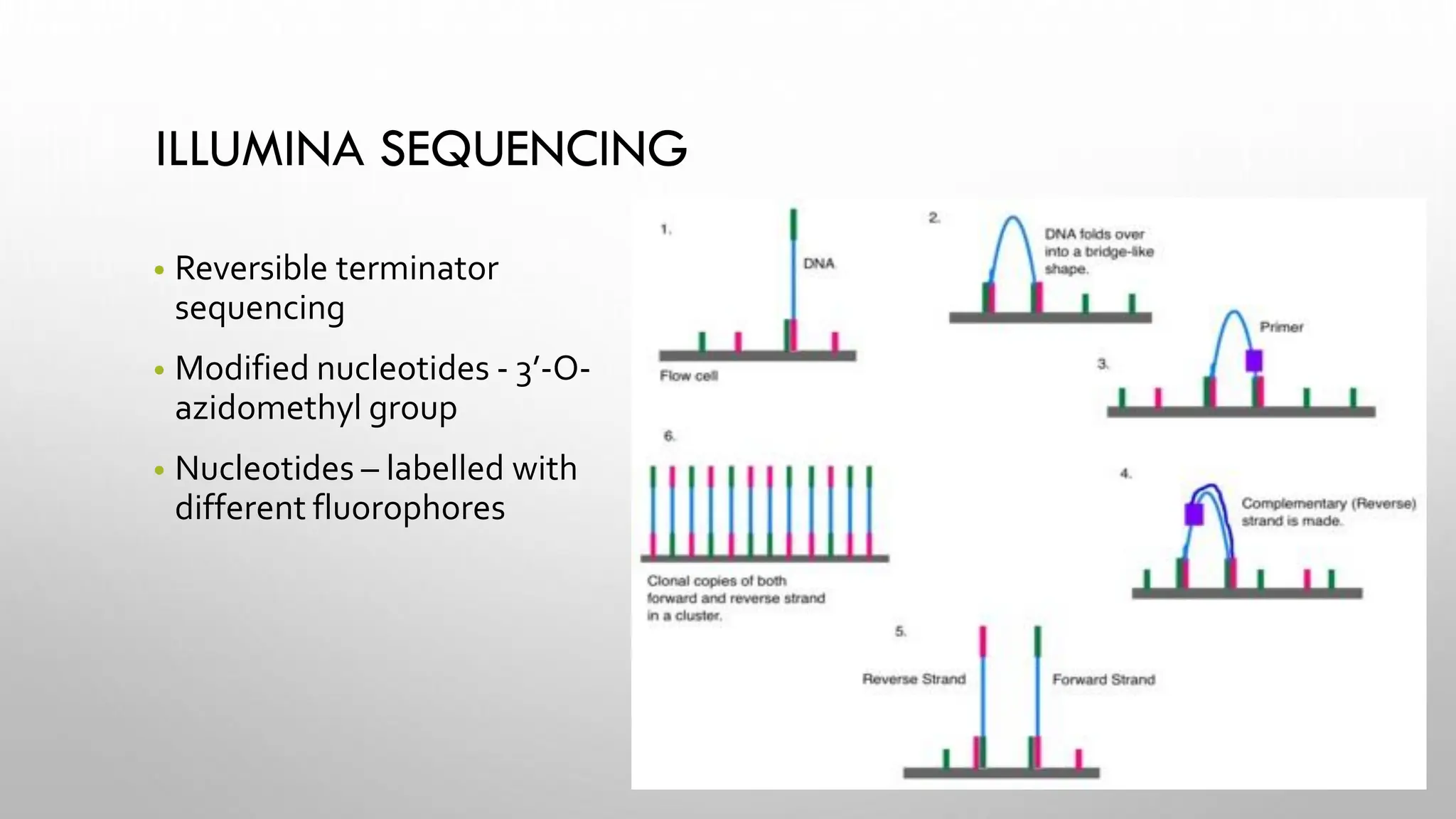
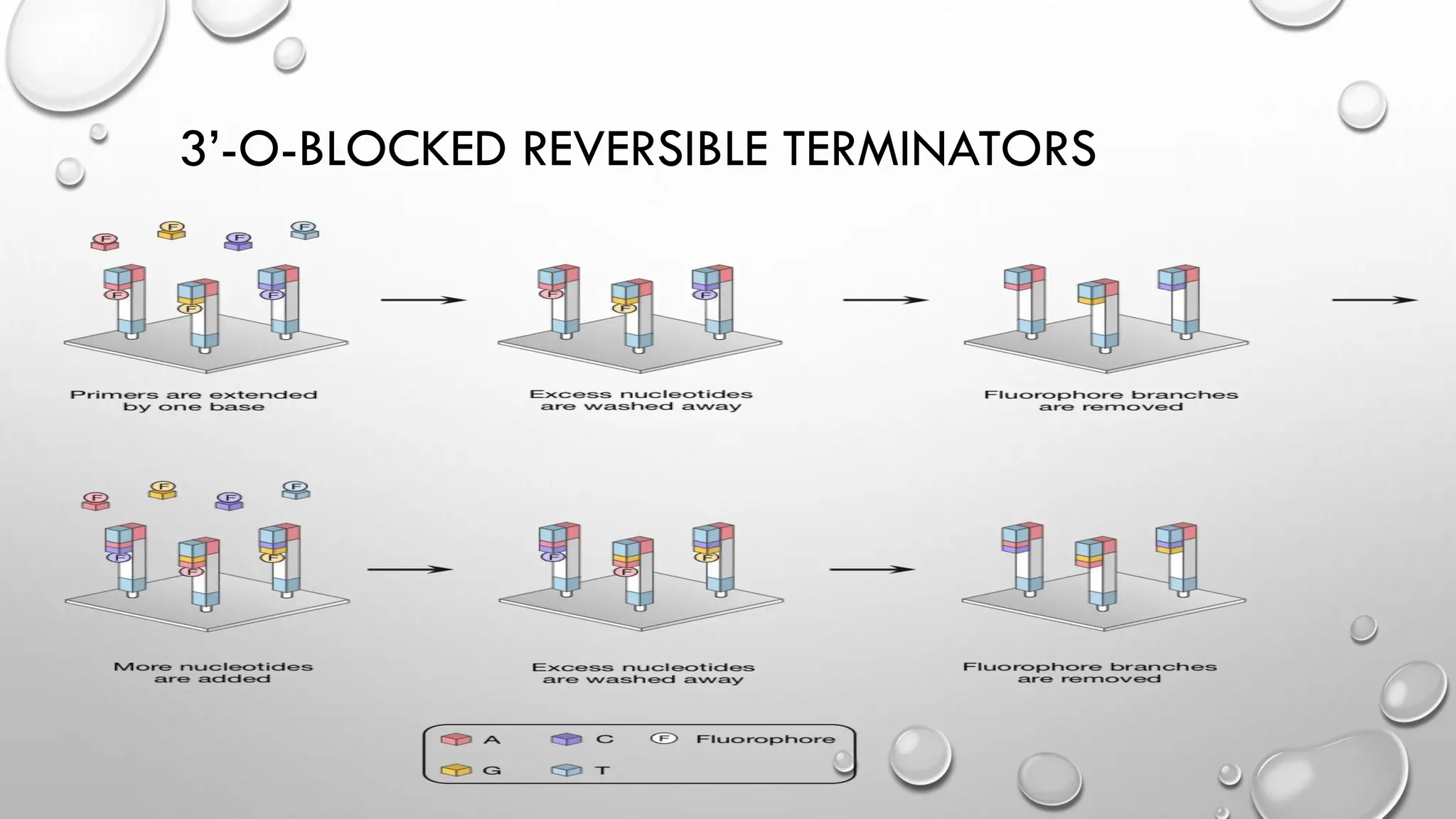
The document provides an overview of nucleic acid blotting techniques, including Southern, Northern, Western, colony, and dot blotting, highlighting their roles in the immobilization and detection of nucleic acids. It also discusses various DNA sequencing methods such as Sanger's chain termination method, Maxam-Gilbert chemical sequencing, and next-generation sequencing techniques, along with their principles and applications. Key processes like sample preparation, sequencing methods, and detection mechanisms are summarized, emphasizing advancements in sequencing efficiency and accuracy.