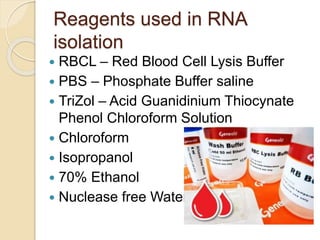
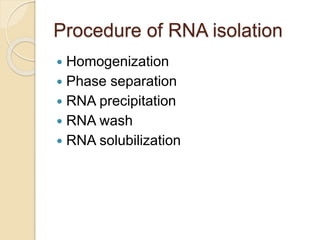
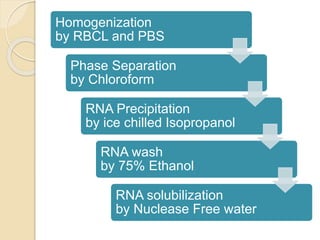
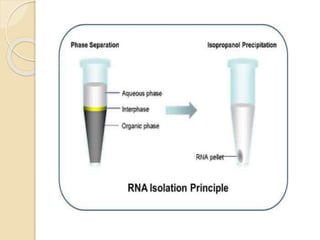
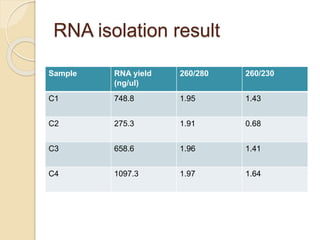
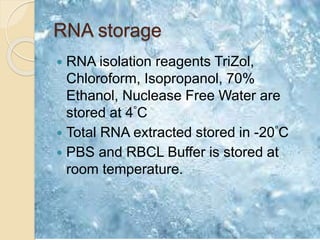
The document discusses DNA and RNA isolation techniques, their methodologies, and applications in various scientific fields. DNA isolation involves extraction from diverse sources and is crucial for forensic analysis, genetic studies, and molecular biology, while RNA isolation emphasizes the need for sensitivity and precision. Both processes include specific reagents, steps for purification, and storage conditions vital for maintaining sample quality.