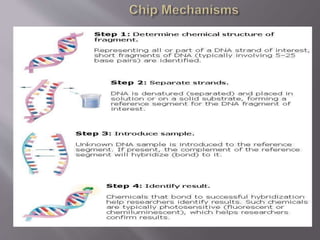
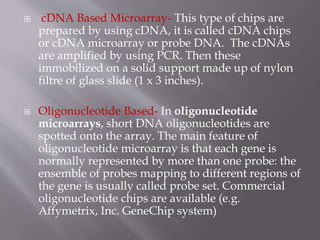
DNA microarrays are solid supports with organized grids of DNA probes that represent genes. Each DNA spot allows comparison of thousands of genes simultaneously. Microarray technology uses DNA chip probes to bind complementary DNA in samples, studying gene expression across entire genomes. Microarrays evolved from Southern blotting and were first used for eukaryotic gene expression profiling in 1995. Microarrays exploit DNA hybridization between nucleotide sequences to screen genomic sequences. They are used for gene expression profiling, drug discovery, diagnostics, and more.