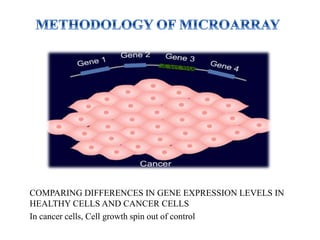
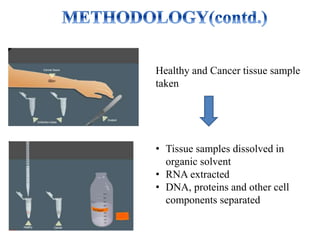
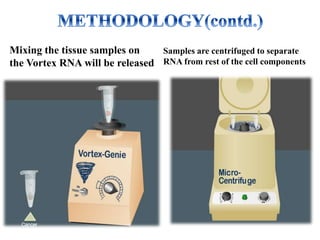
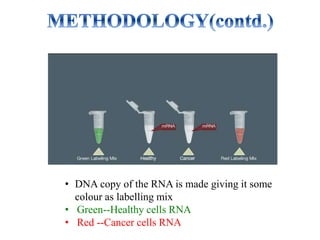
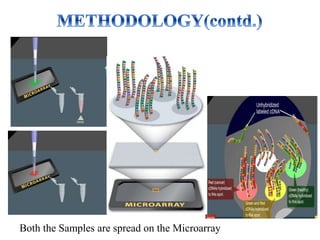
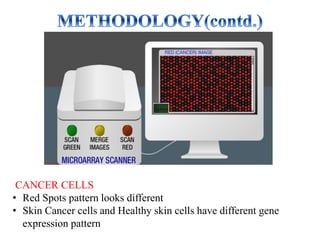
DNA microarrays, also known as DNA chips, are solid supports with DNA attached in an organized grid pattern. Each spot represents a single gene. Microarray technology allows screening of 1,00,000 genomic or cDNA sequences for presence in a single hybridization. RNA from healthy and cancer tissue samples are labeled with different colors, applied to a microarray, and analyzed to see which genes are expressed differently between healthy and cancer cells. DNA microarrays have many applications and provide large amounts of gene expression data quickly, but are expensive to create and results require complex analysis.


![3
Introduction:
Definition: DNA microarrays are solid supports, usually of glass or
silicon, upon which DNA is attached in an organized grid fashion.
Each spot of DNA, called a probe, represents a single gene.
There are several synonyms of DNA microarrays such as DNA
chips, gene chips, DNA arrays, gene arrays and biochips.
[Ref: Presscot (Book for Microbiology), www.wikipedia.org]
History: Microarray technology evolved from Southern blotting,
where fragmented DNA is attached to a substrate and then probed with
a known DNA sequence.
The use of miniaturized microarrays for gene expression profiling was
first reported in 1995, and a complete eukaryotic genome
(Saccharomyces cerevisiae) on a microarray was published in 1997.](https://image.slidesharecdn.com/dnamicroarray-180209113652/85/Dna-microarray-3-320.jpg)






![20
• Provides data for thousands of genes.
• One experiment instead of many.
• Fast and easy to obtain results.
• Huge step closer to discovering cures for diseases and cancer.
• Different parts of DNA can be used to study gene expresion.
ADVANTAGES
[Ref: www.biotechnologyforums.com, www.ehow.com]](https://image.slidesharecdn.com/dnamicroarray-180209113652/85/Dna-microarray-20-320.jpg)

