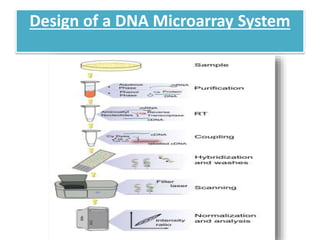
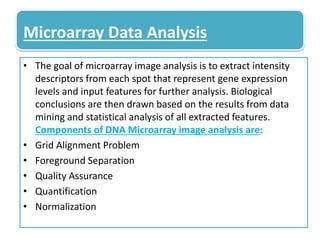
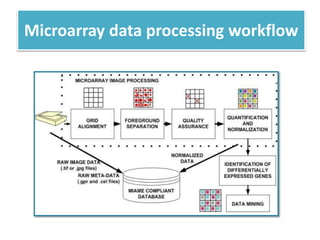
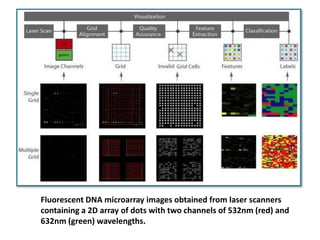
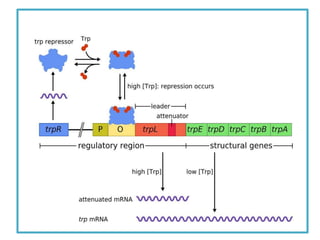
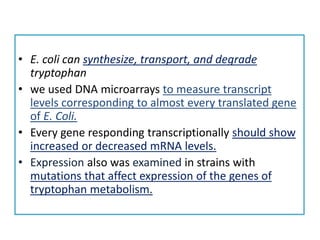
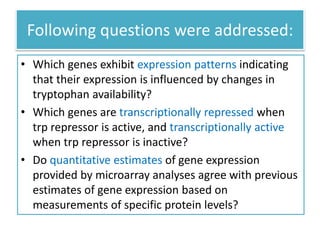
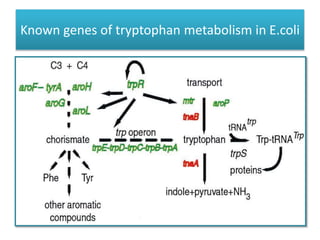
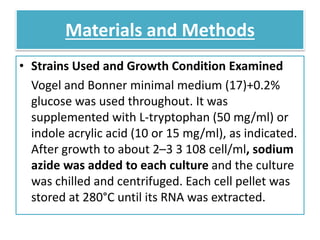
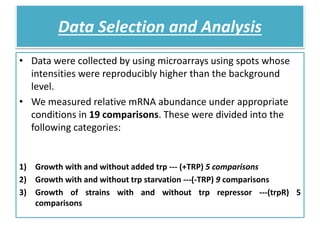
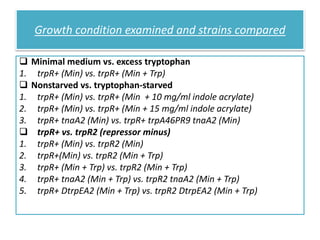
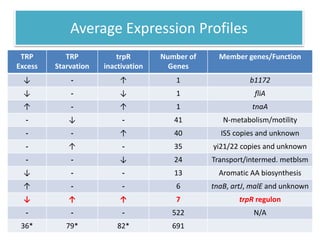
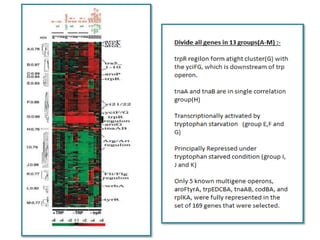
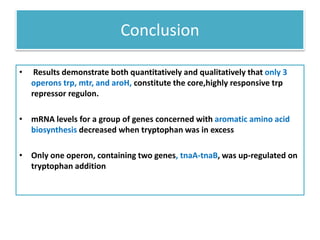
This document describes a study that used DNA microarrays to analyze changes in gene expression related to tryptophan metabolism in E. coli under different physiological conditions and genetic mutations. The study identified genes whose expression levels changed with tryptophan availability, tryptophan starvation, and inactivation of the tryptophan repressor. Only a small core set of operons including trp, mtr and aroH showed highly responsive changes in expression levels. mRNA levels for aromatic amino acid biosynthesis genes decreased with excess tryptophan, while only the tnaA-tnaB operon increased. The results provide quantitative validation of genes known to be involved in tryptophan metabolism.