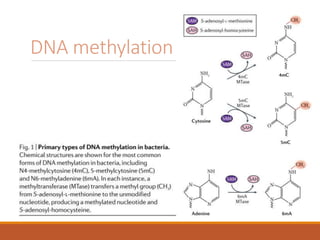
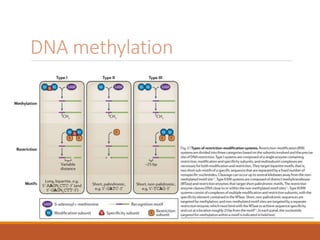
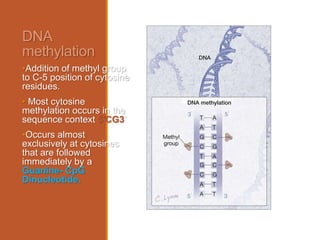
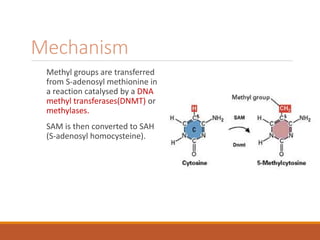
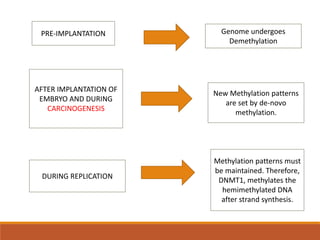
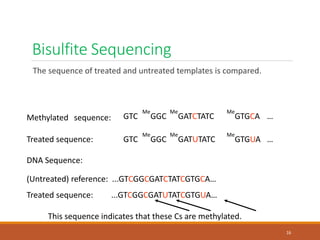
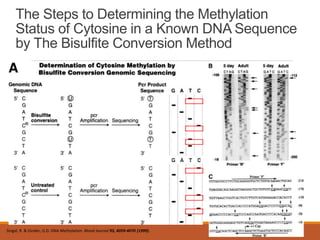
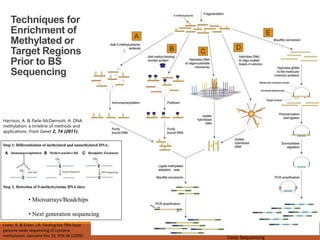
This document discusses DNA methylation. It begins by explaining that DNA methylation is an epigenetic mechanism where a methyl group is added to cytosine residues in DNA, without changing the DNA sequence. This modification regulates gene expression and is involved in cell differentiation. The document then focuses on DNA methylation in more detail, describing where in the genome it occurs, the enzymes involved like DNMTs, and techniques to detect methylation like bisulfite sequencing. It explains that DNA methylation patterns are important for normal cell functions and development.