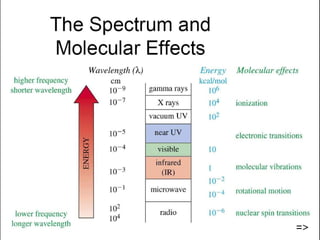
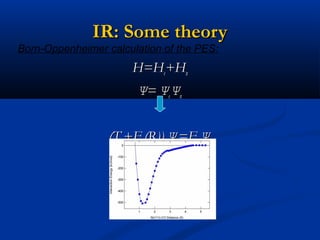
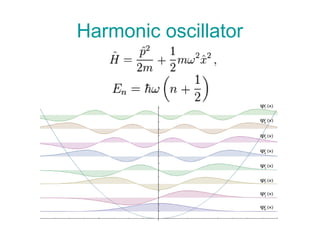
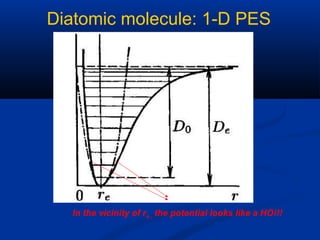
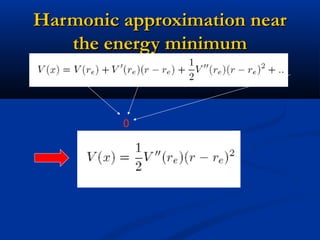
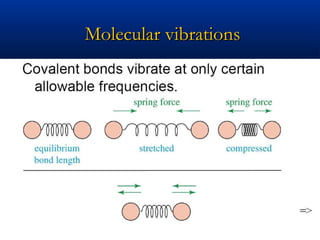
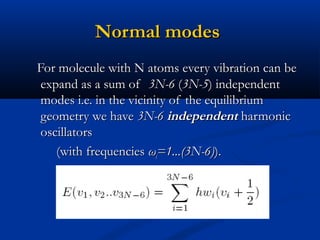
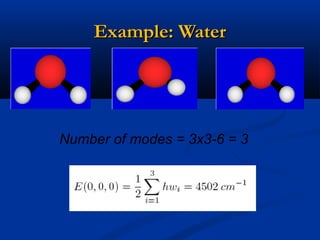
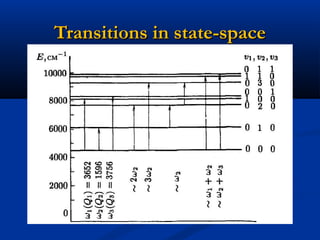
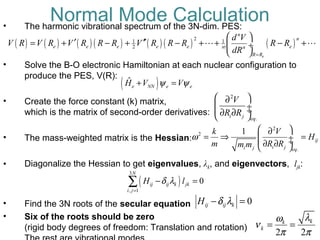
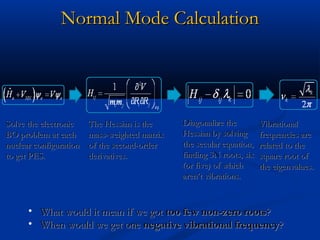
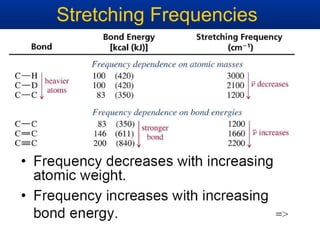
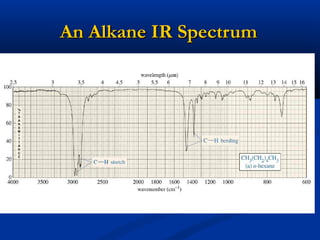
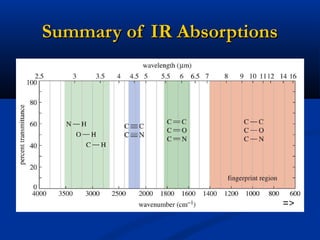
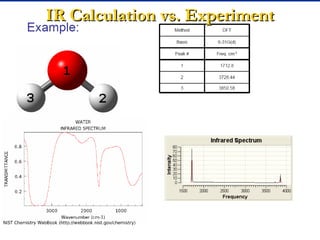
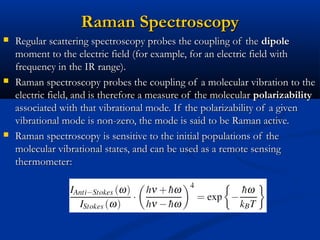
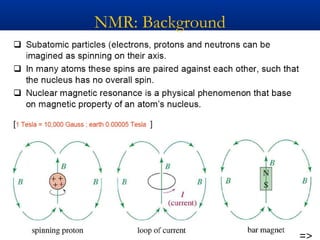
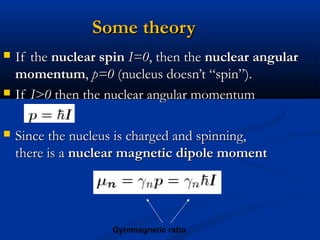
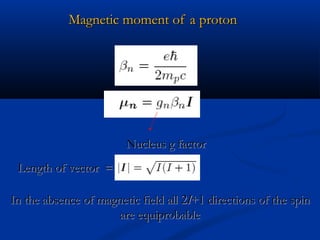
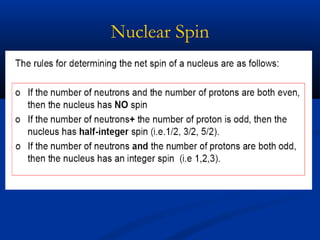
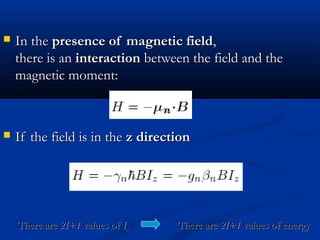
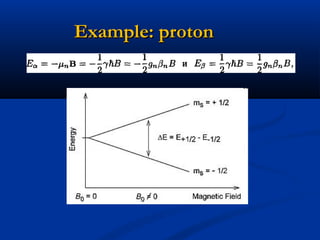
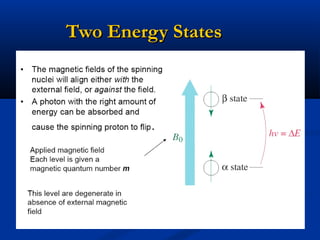
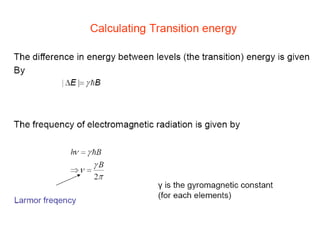
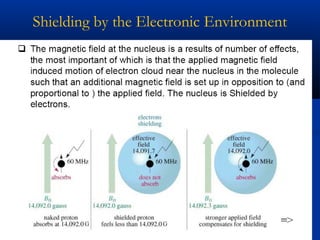
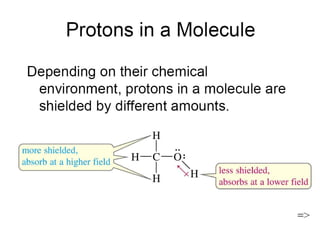
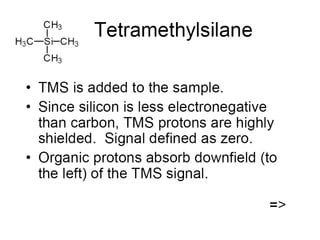
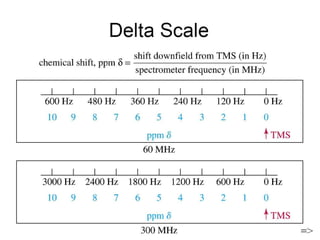
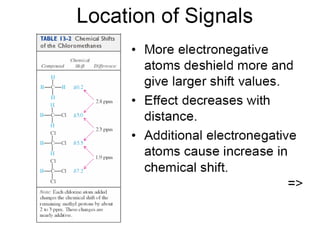
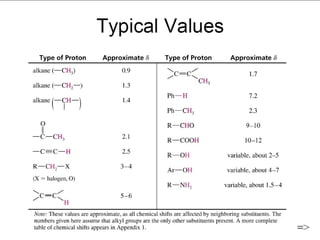
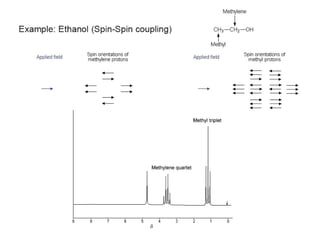
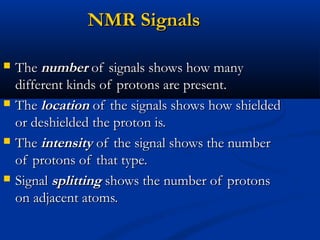
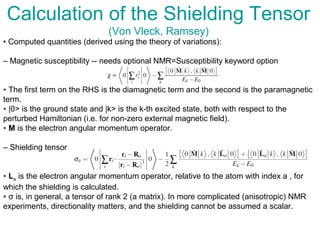
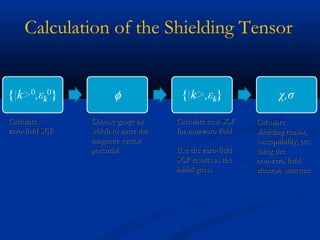
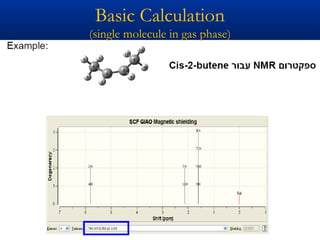
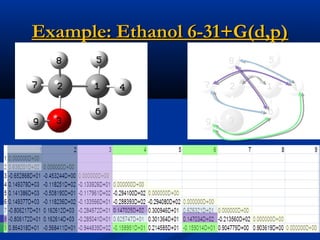
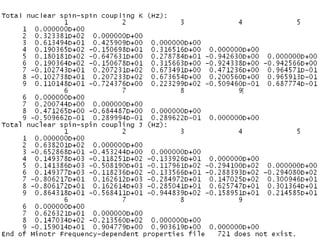
The document outlines the principles and calculations involved in infrared (IR) and nuclear magnetic resonance (NMR) spectroscopy, including their theoretical foundations and practical applications. It discusses the methodology for calculating vibrational frequencies, the role of electronic environment in NMR, as well as limitations and techniques like Raman spectroscopy. The document also emphasizes the uniqueness of molecular spectra and how they can confirm molecular structures.