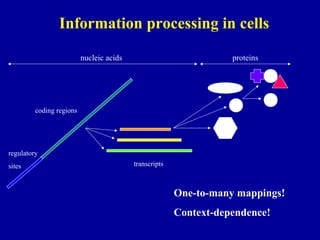
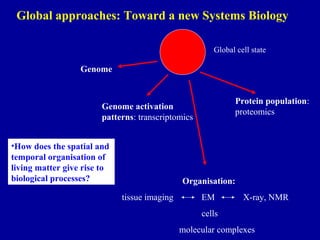
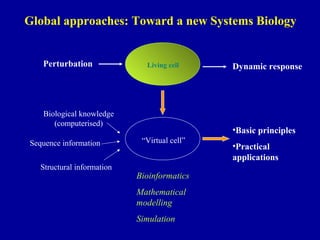
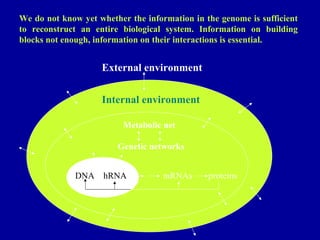
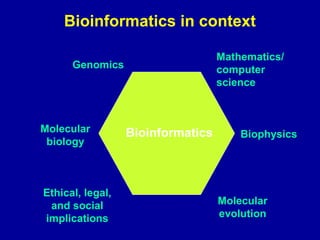
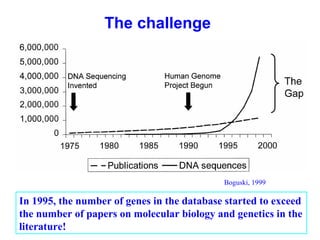
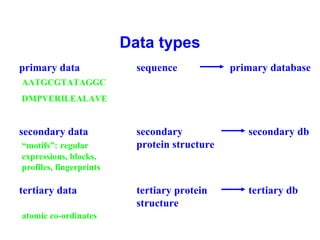
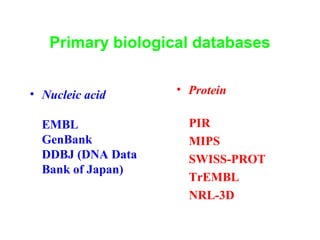
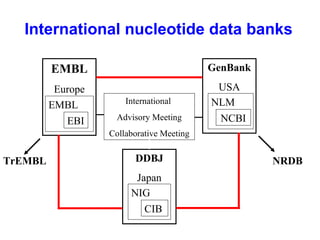
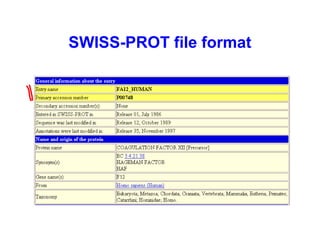
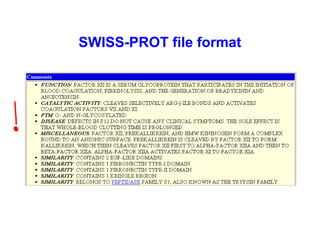
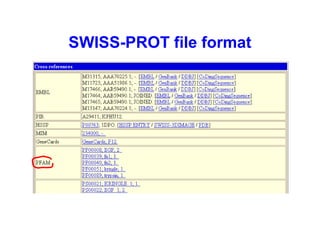
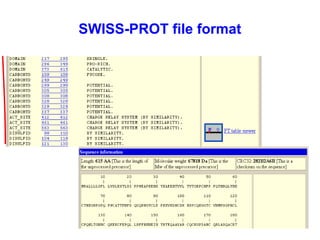
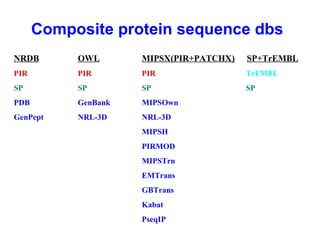
Bioinformatics is an interdisciplinary field that uses computational tools to analyze and manage biological data such as genes, genomes, proteins, and medical information. It involves developing mathematical models to understand relationships in complex biological systems. Key areas include analyzing protein and gene sequences, structures, and functions; understanding evolution and molecular interactions; and developing "virtual cells" through integrated modeling. Major challenges include integrating heterogeneous biological data sources and developing robust computational methods.