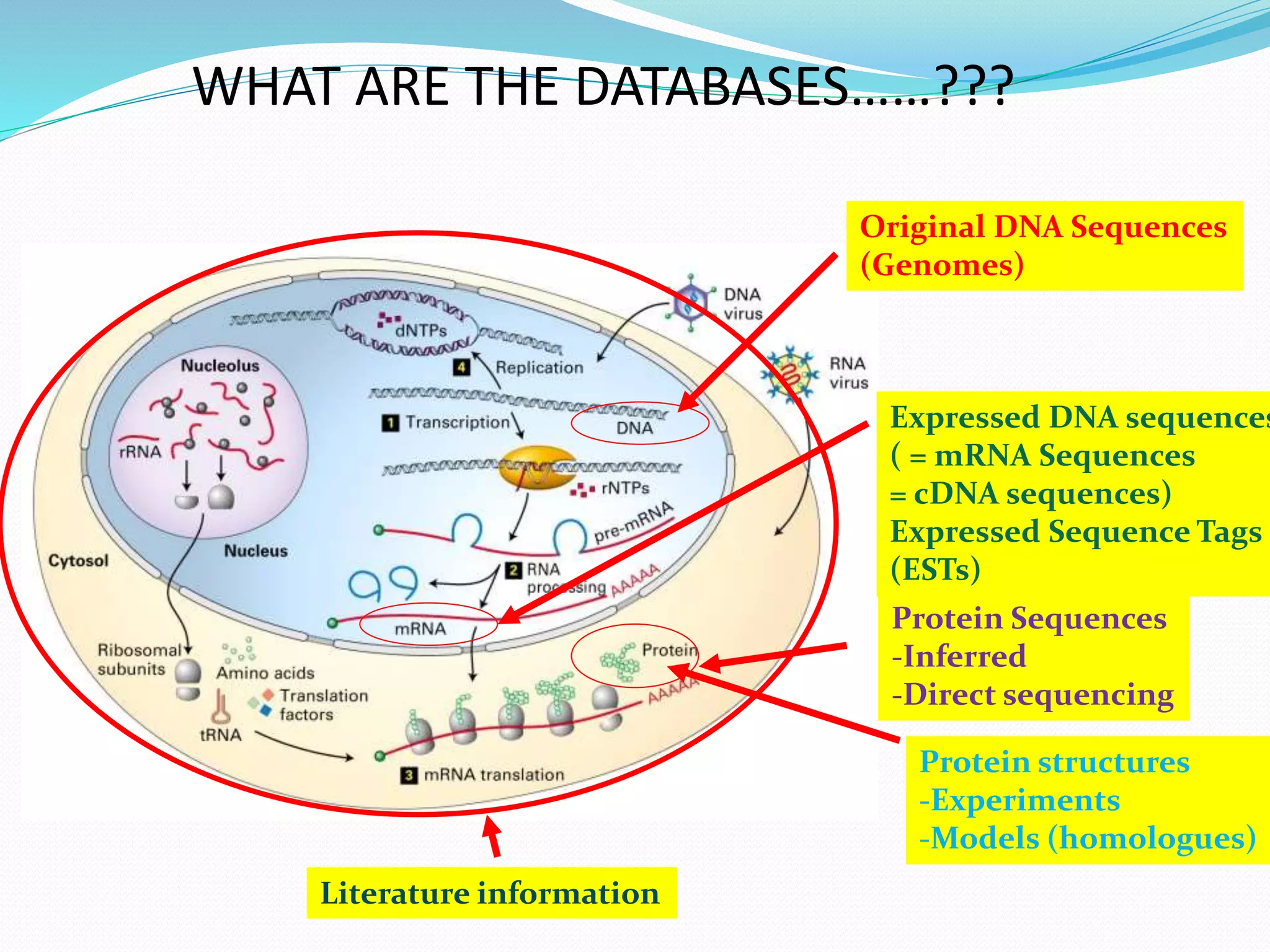
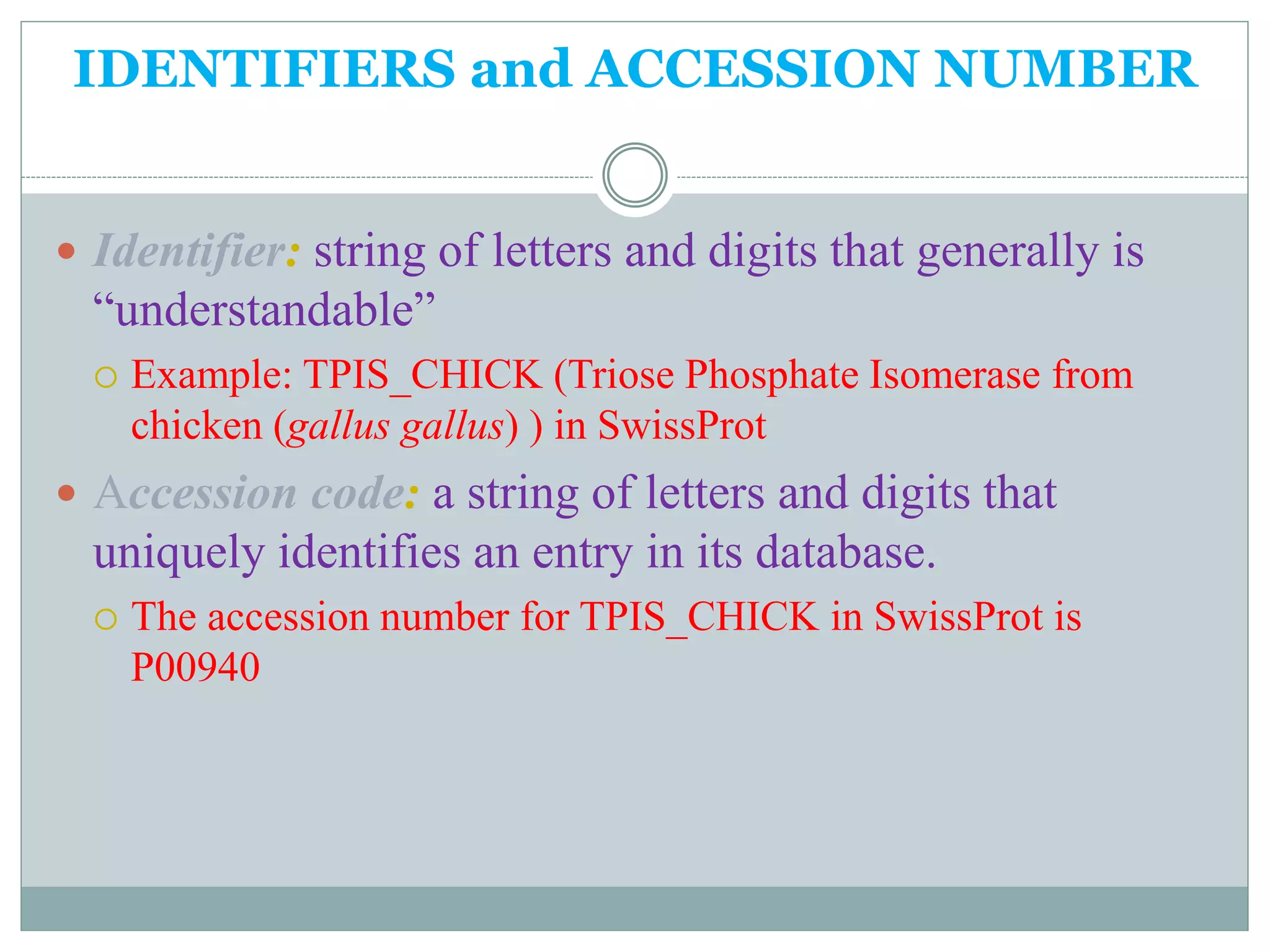
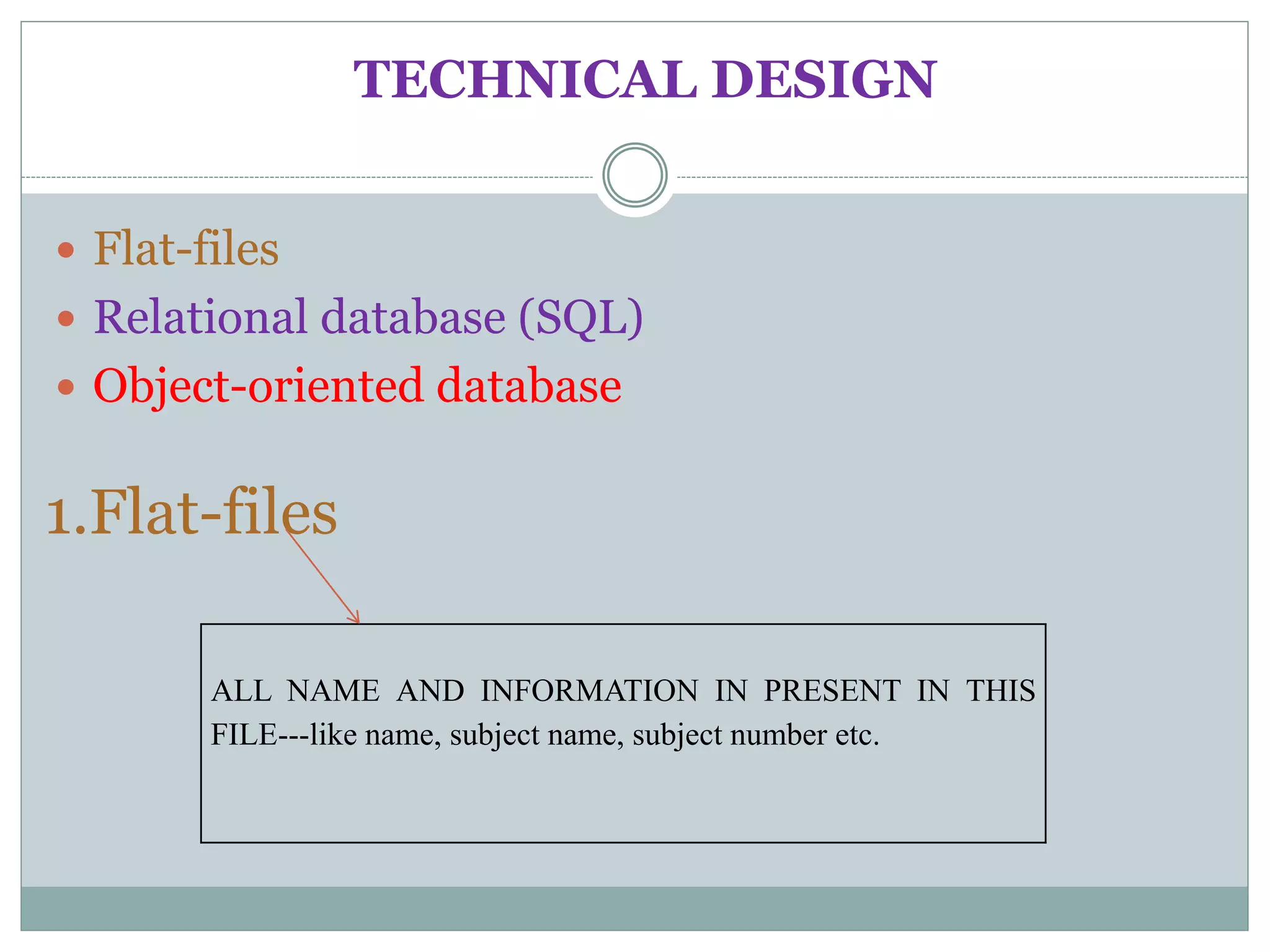
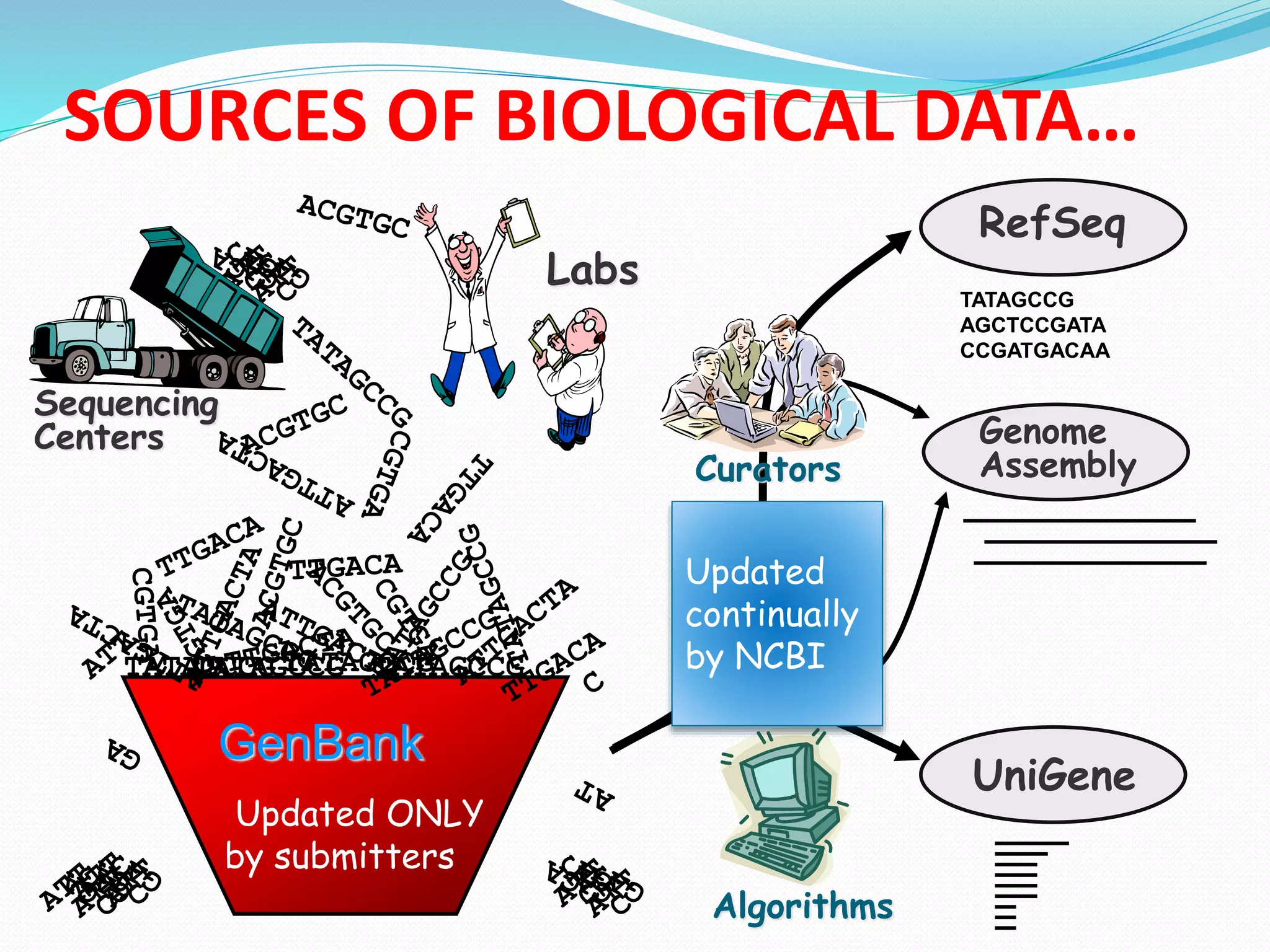
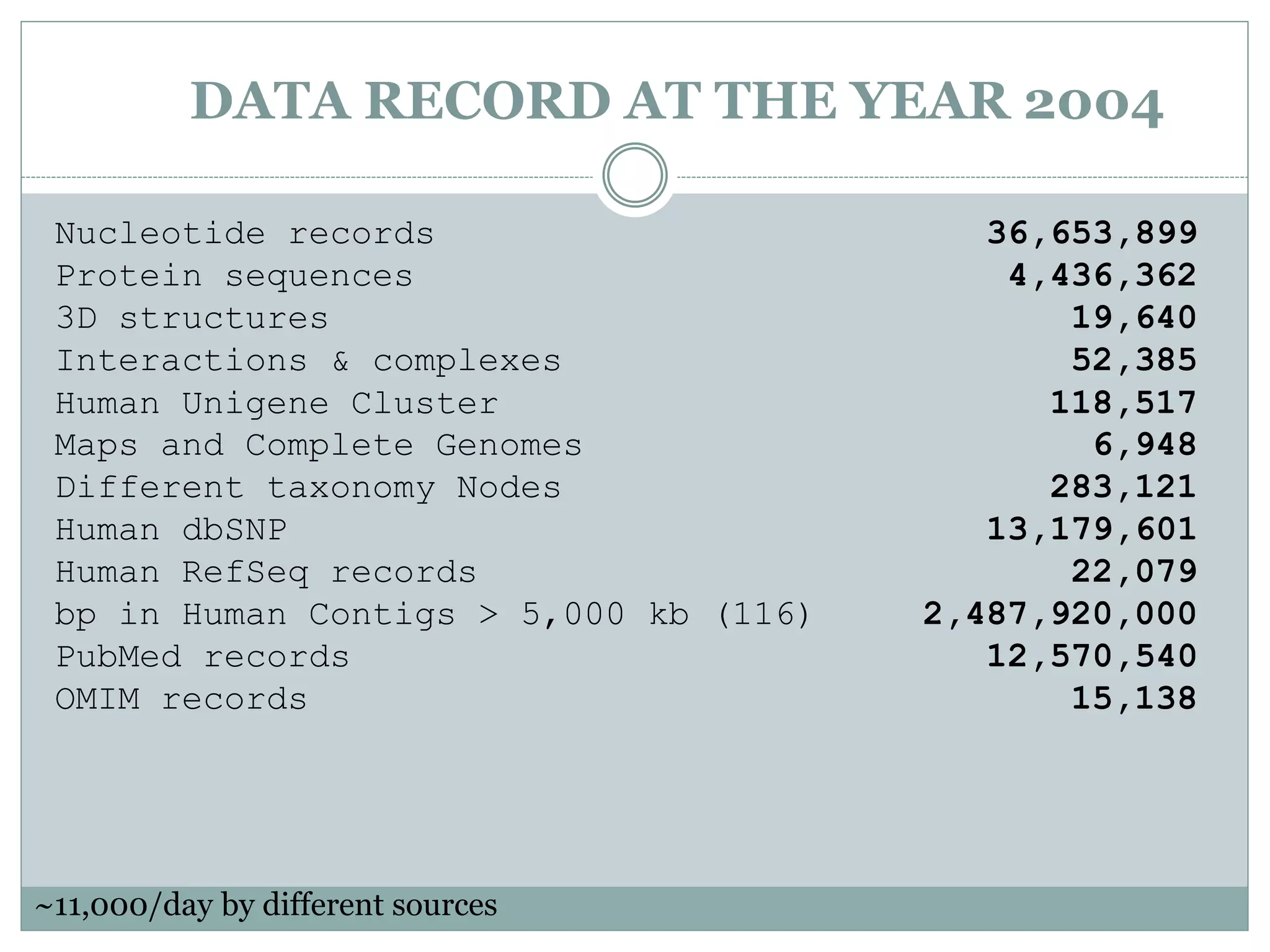
The document discusses biological databases, their history, and significance in life sciences, detailing types such as protein structures and gene expression databases. It covers database design, maintenance, and applications for scientific research, emphasizing data integrity and access. The historical context includes milestones in database development and highlights the importance of these databases in consolidating biological data.