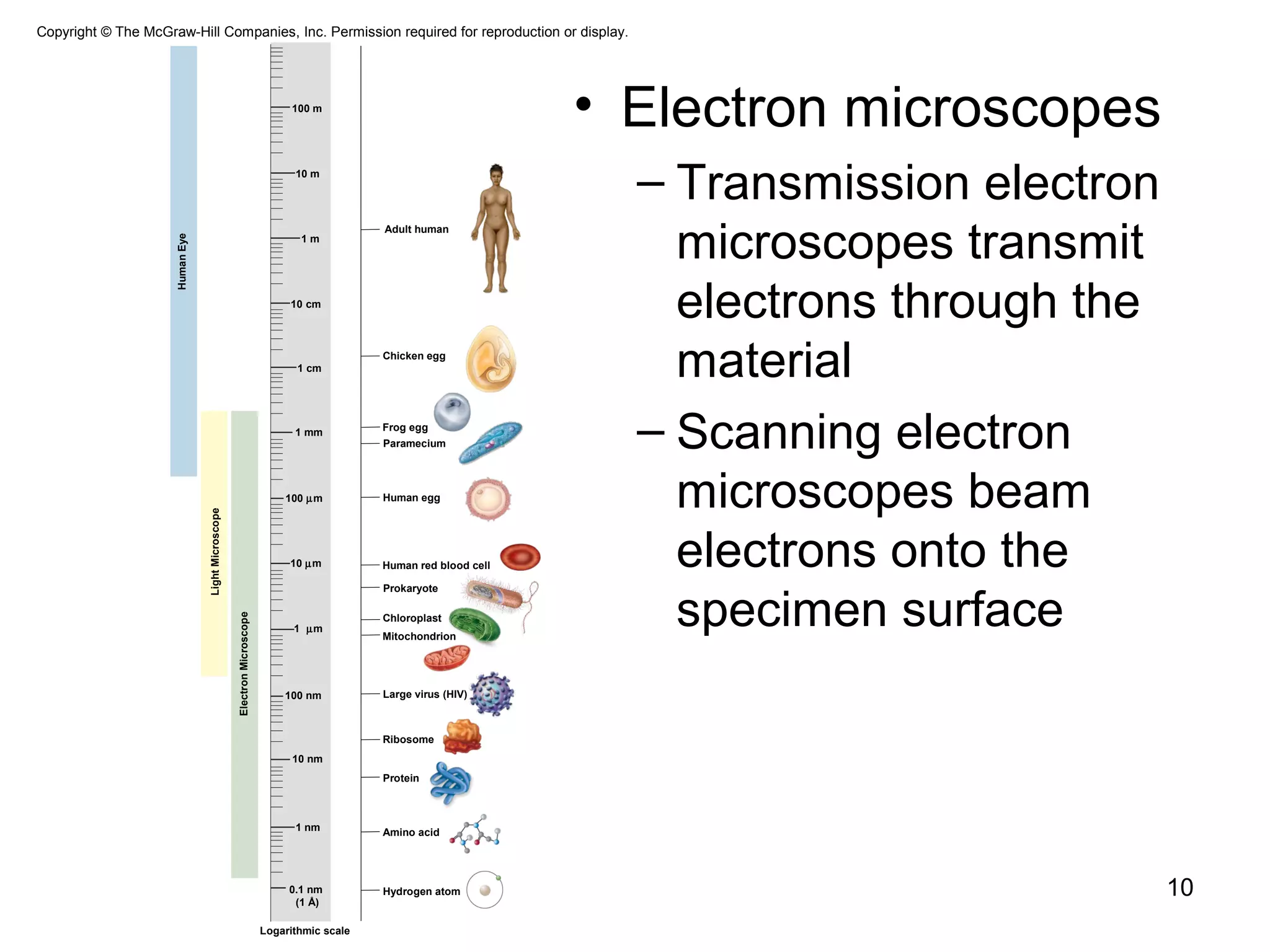
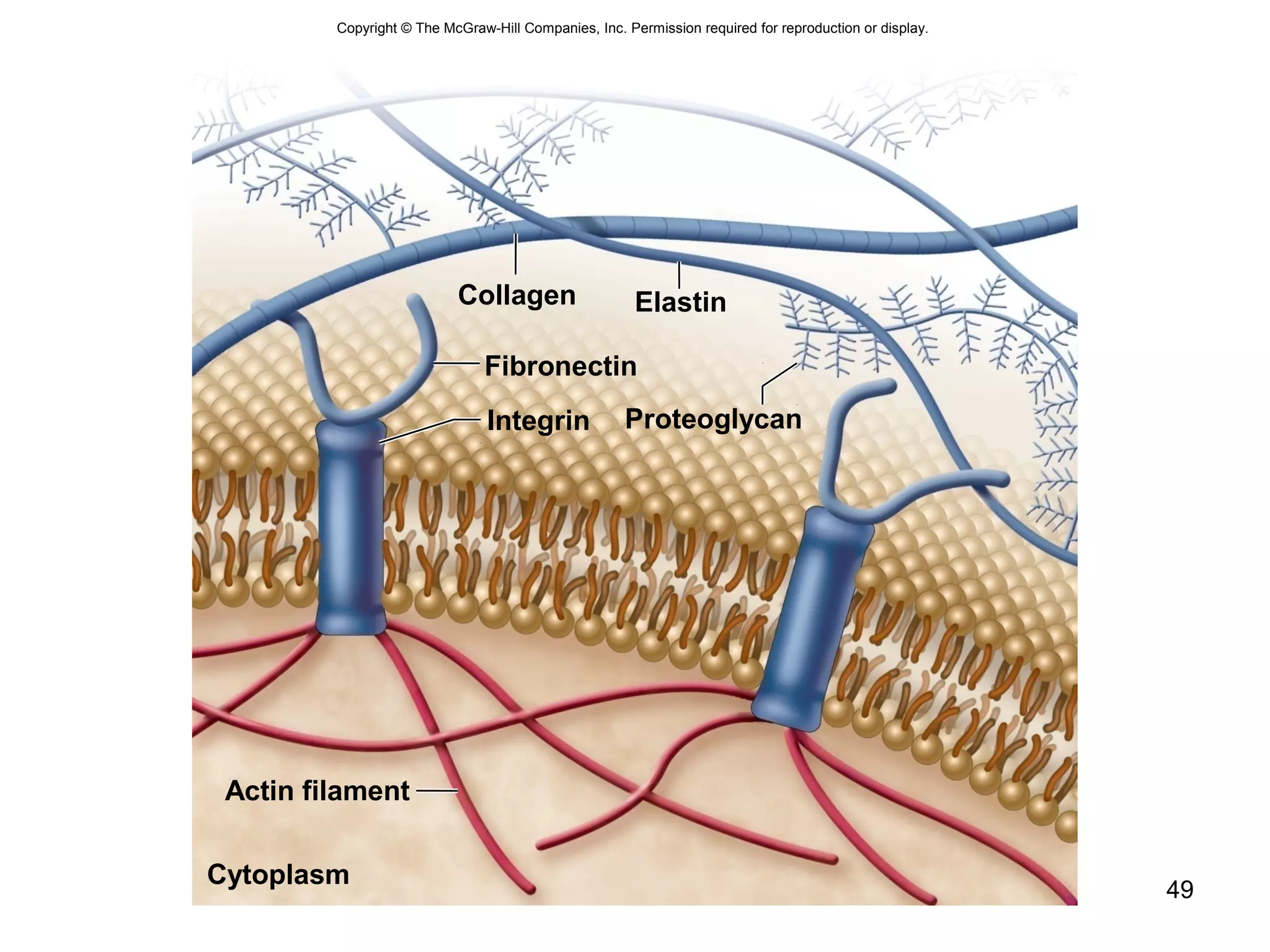
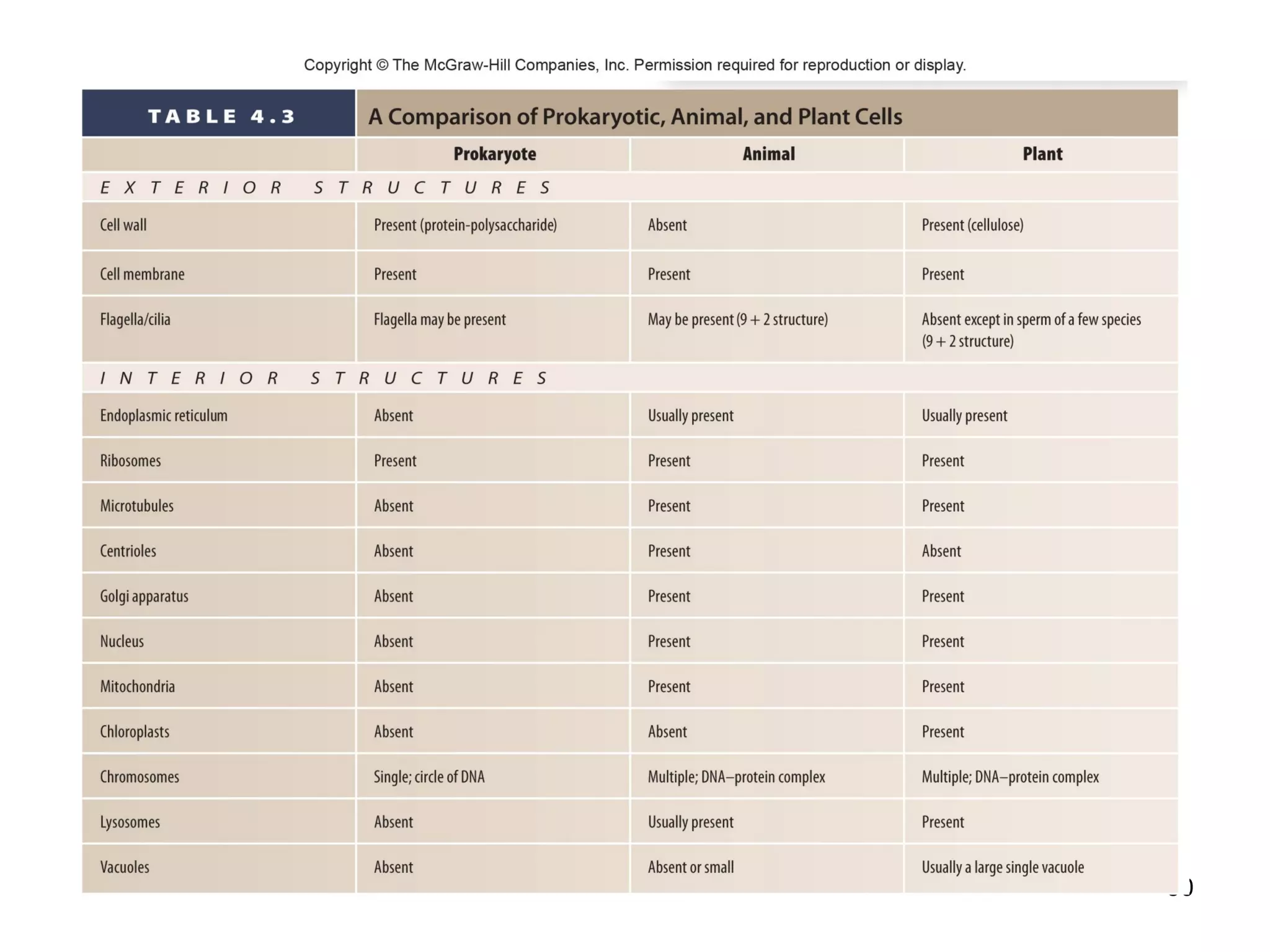
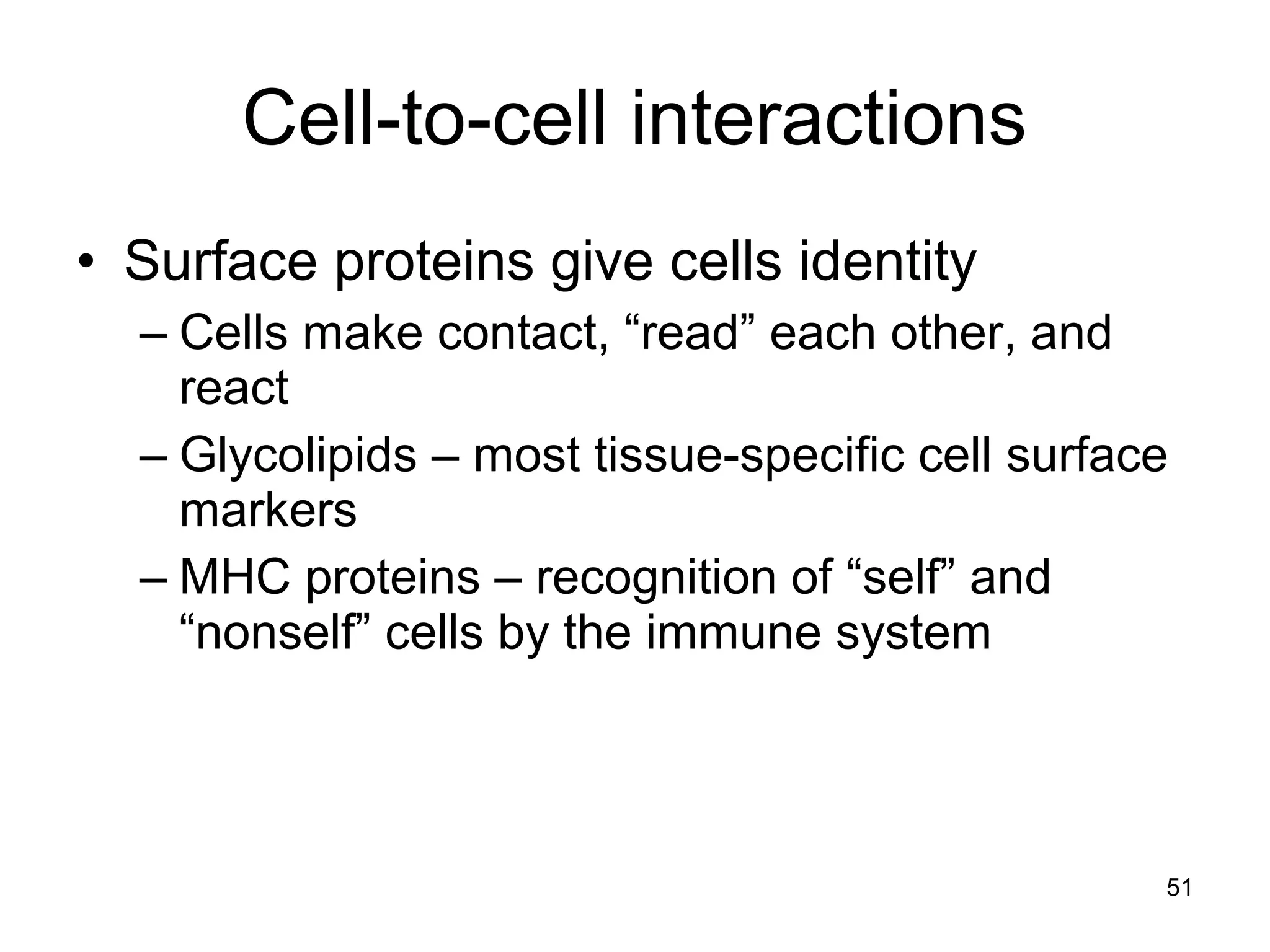
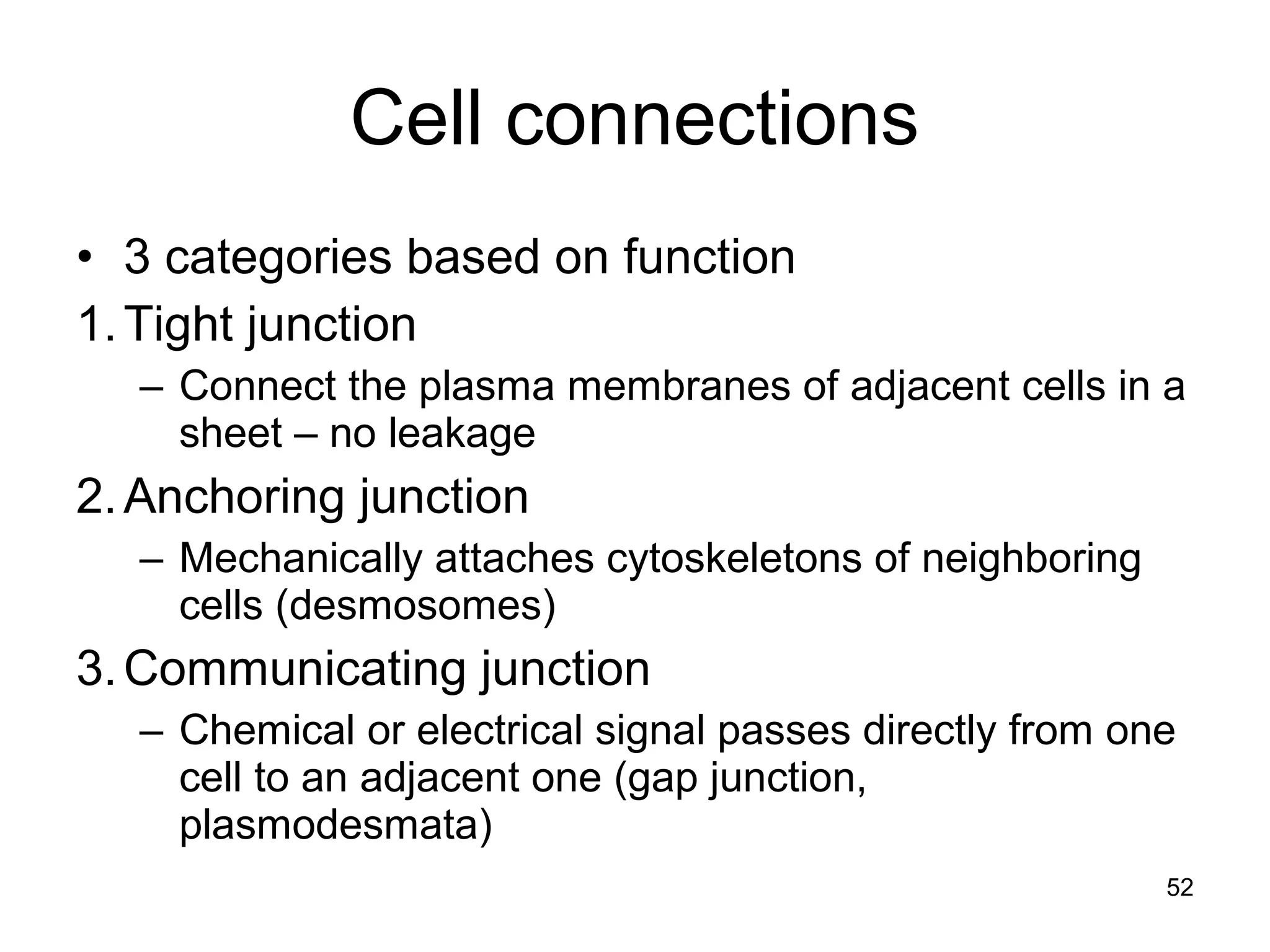
1. The document outlines key aspects of cell structure, including the discovery of cells and the cell theory. It describes the basic components of prokaryotic and eukaryotic cells.
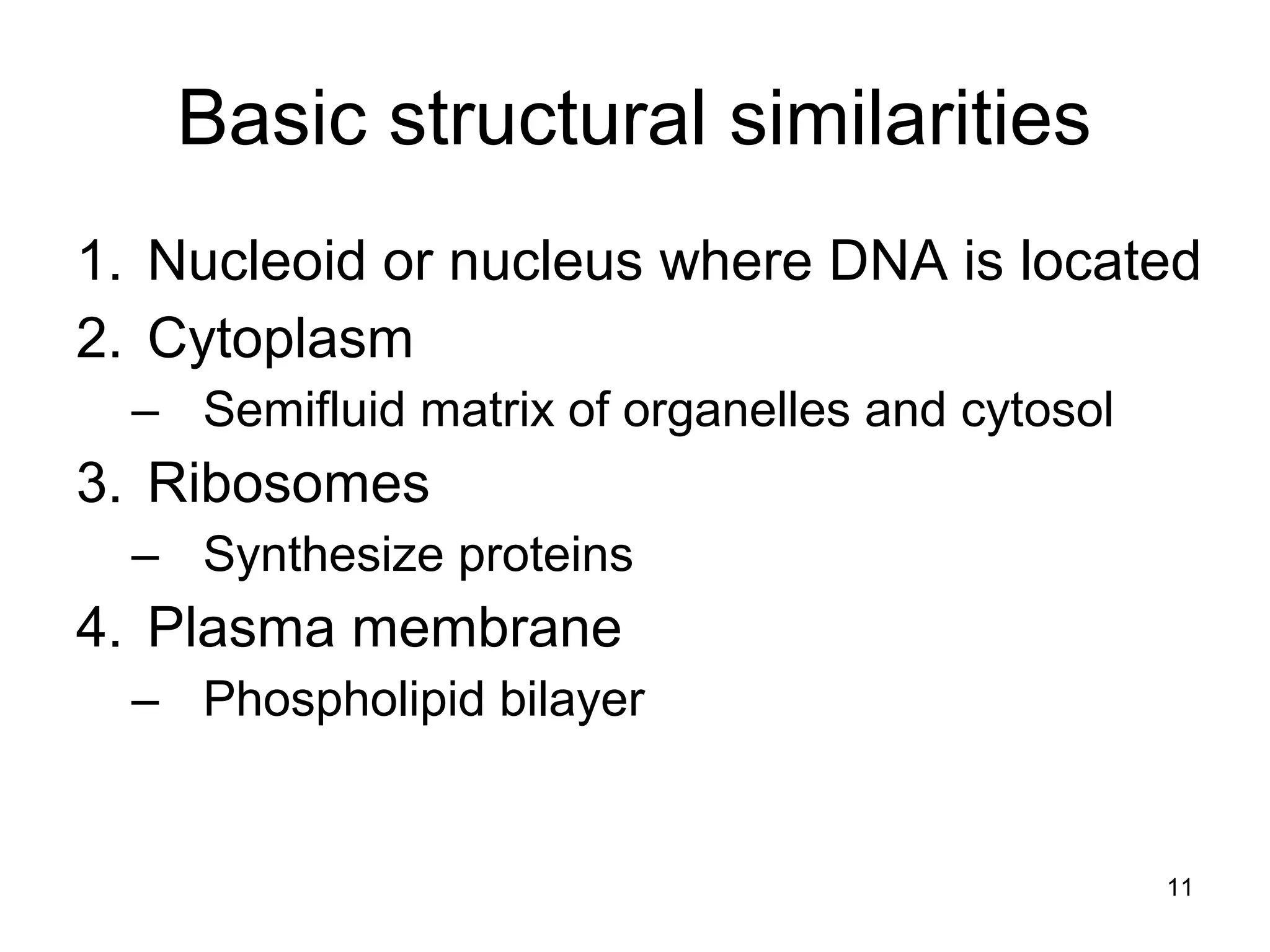
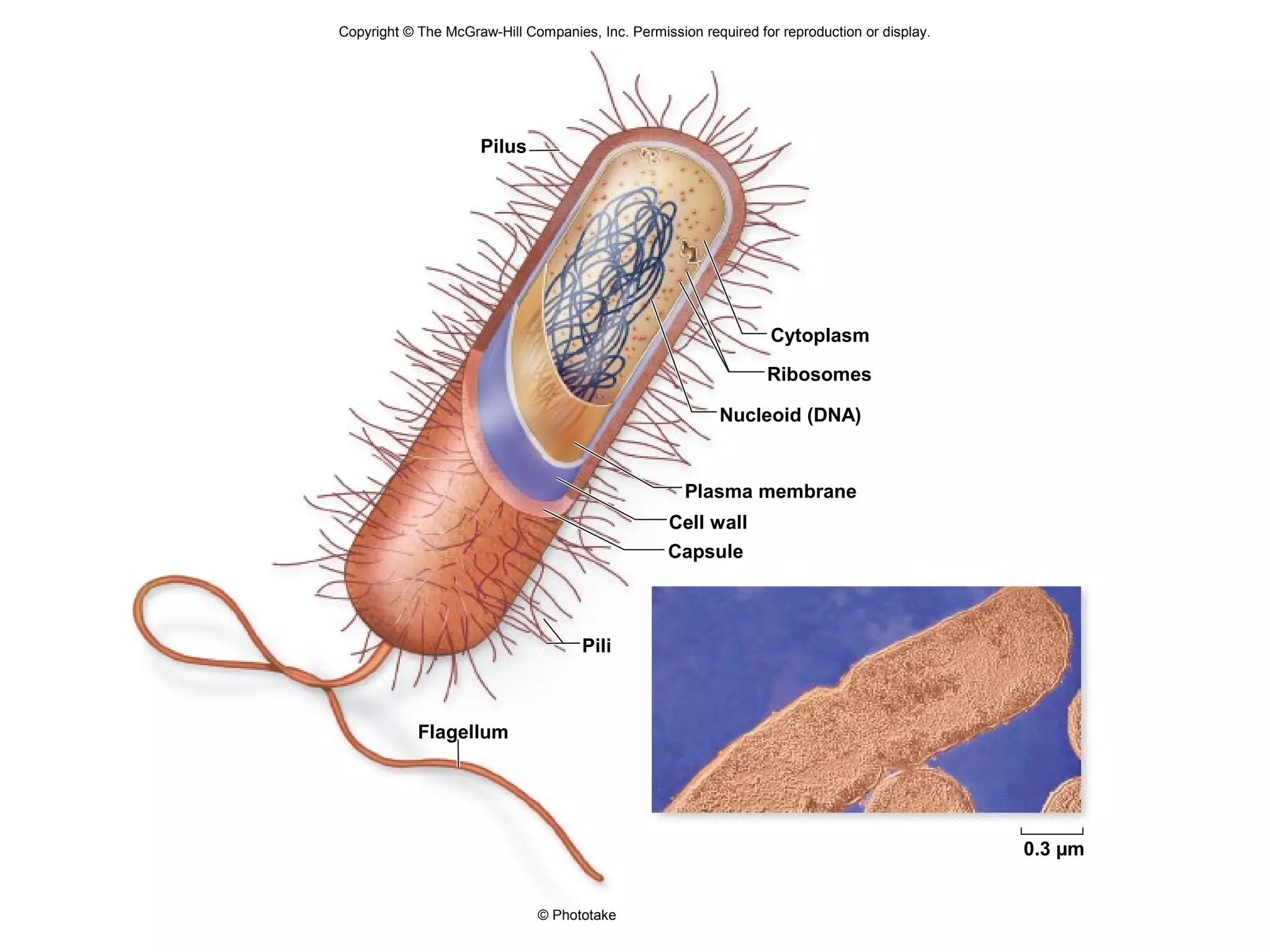
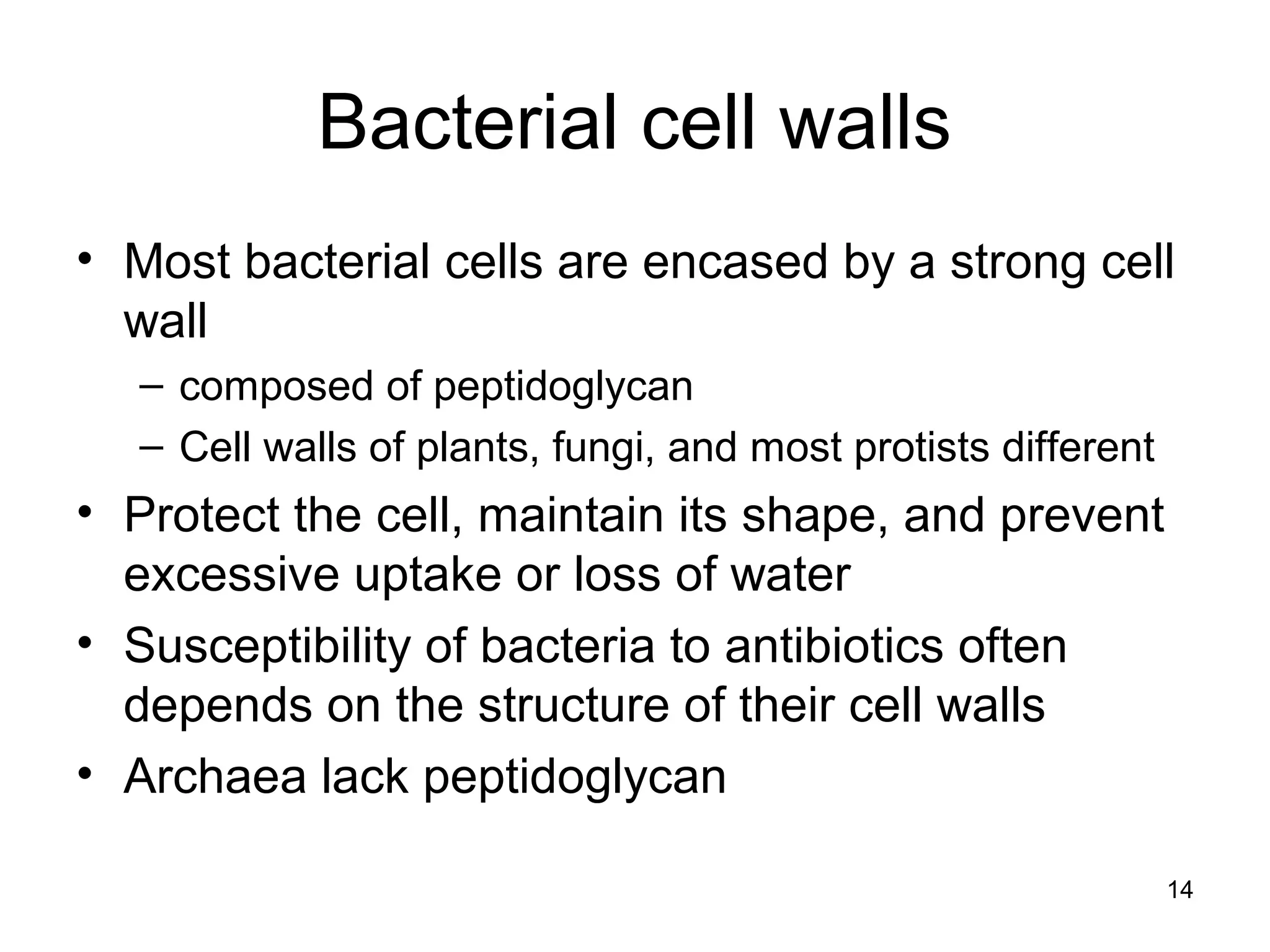
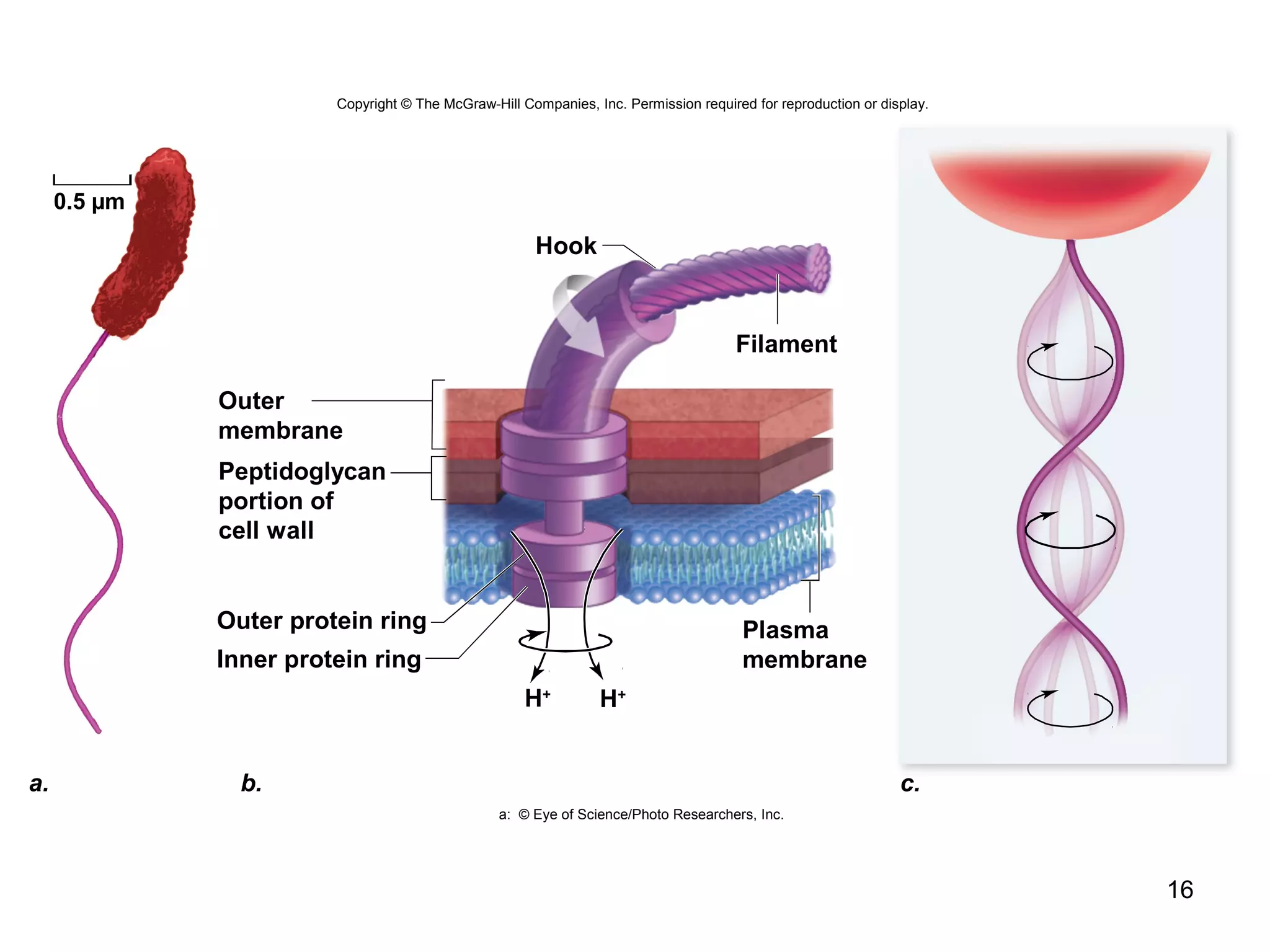
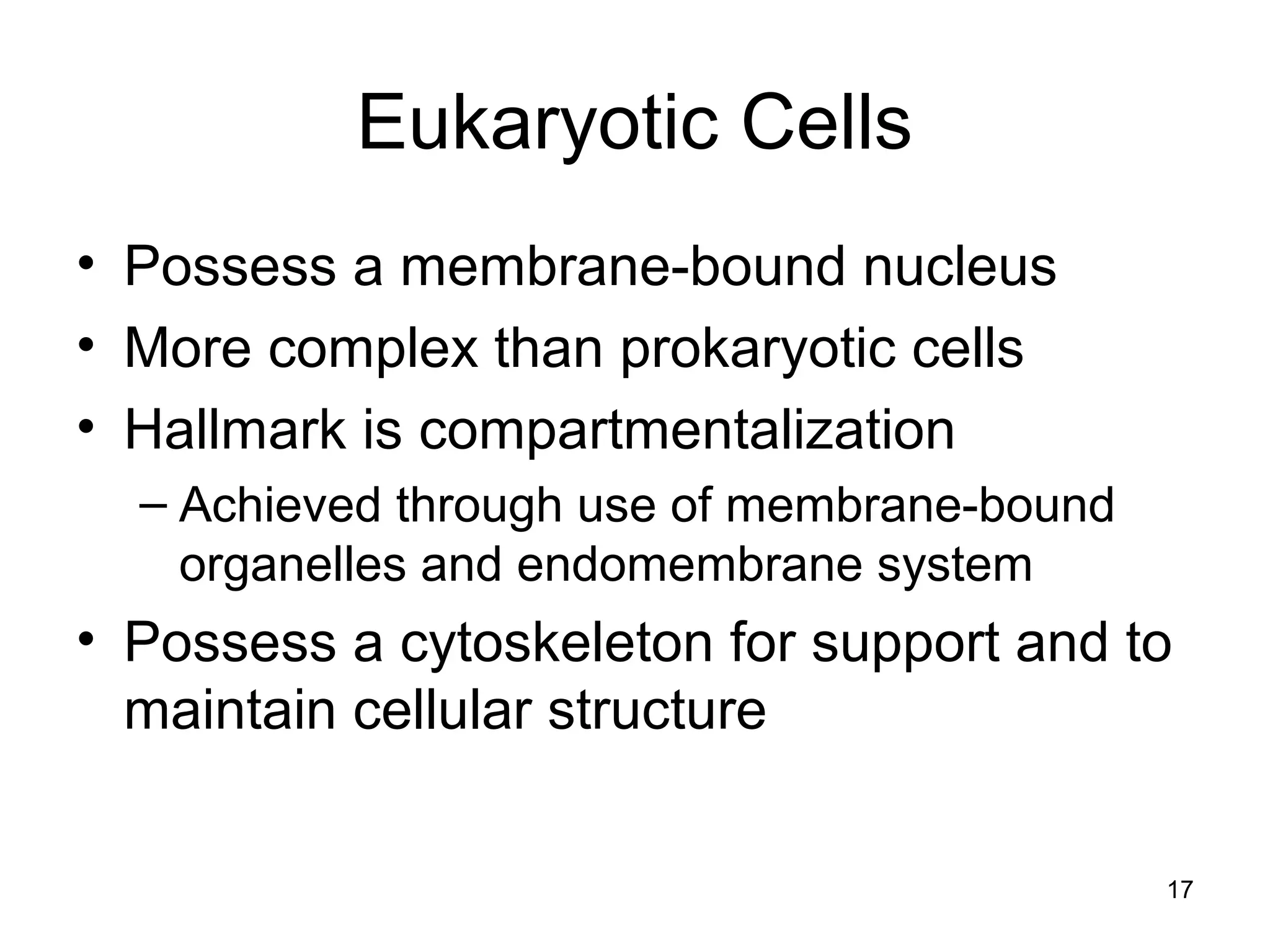
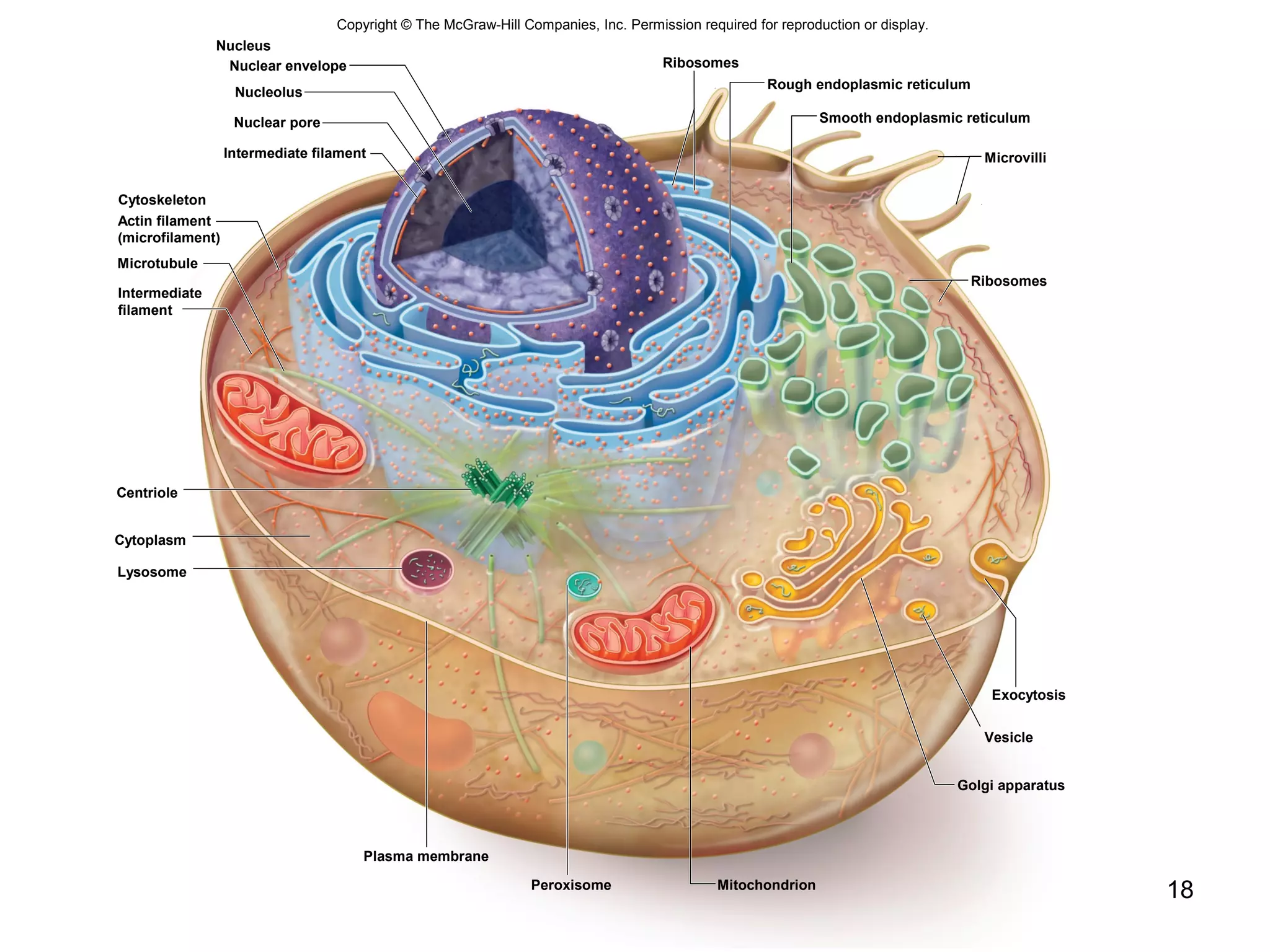
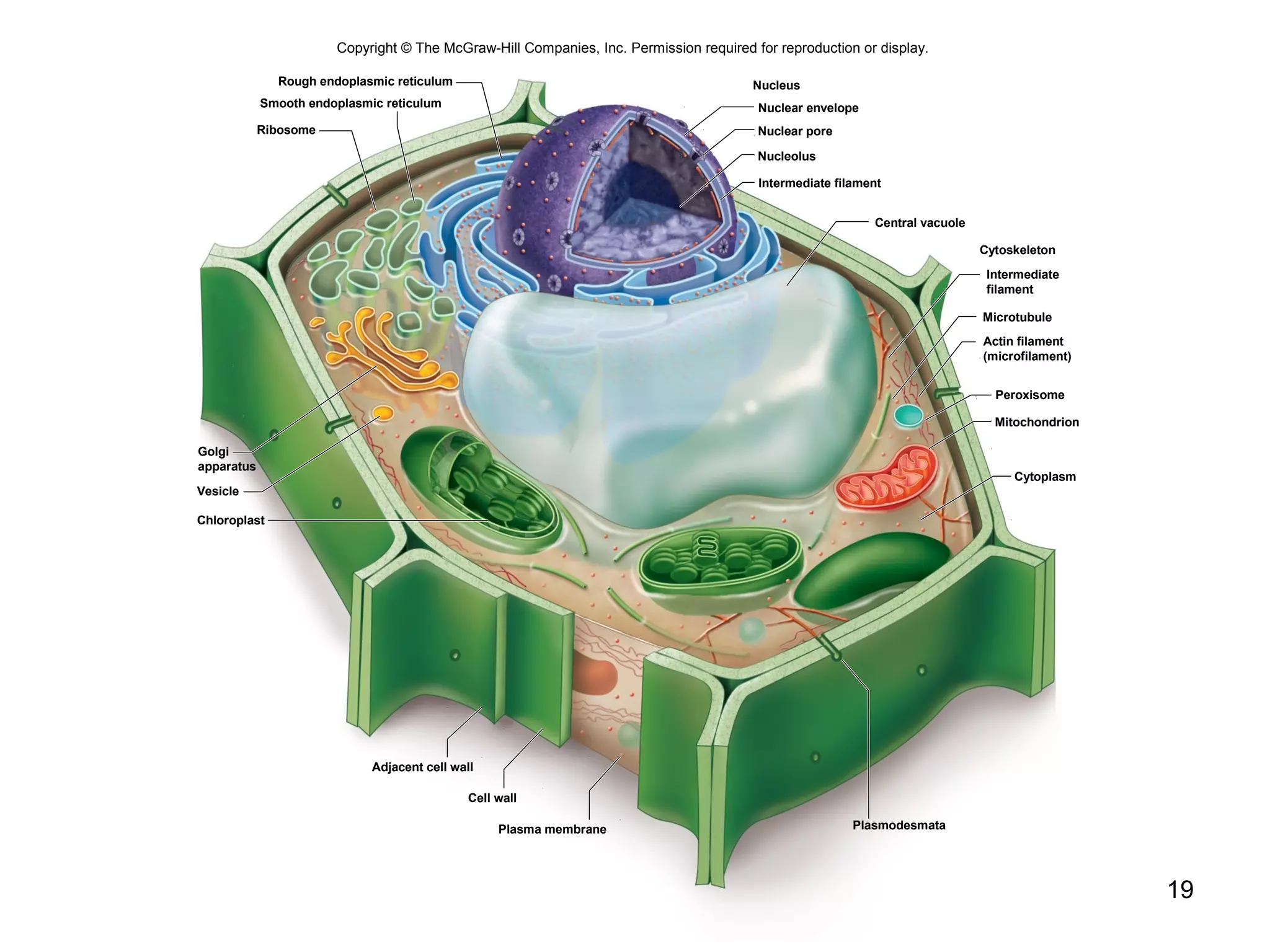
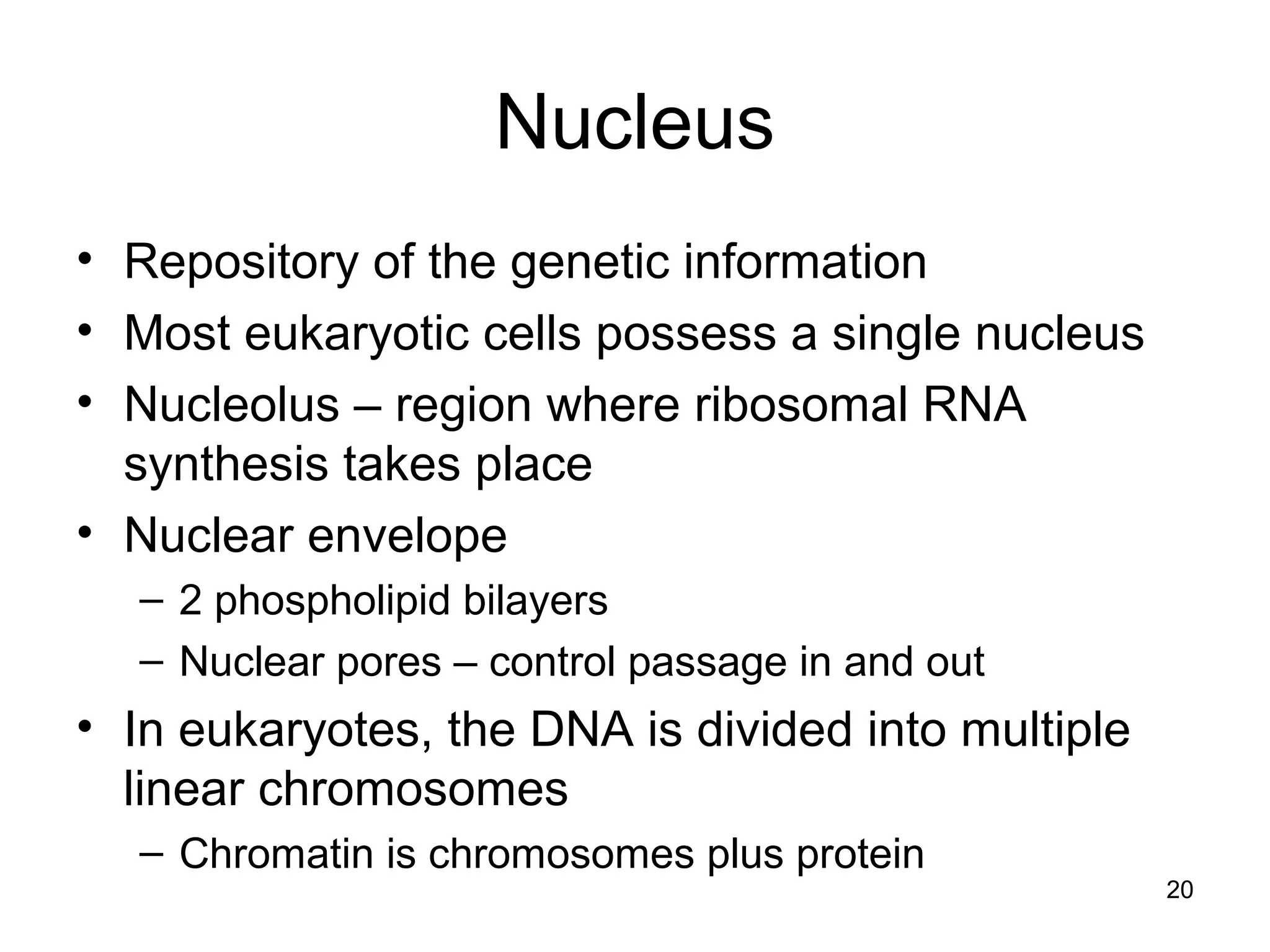
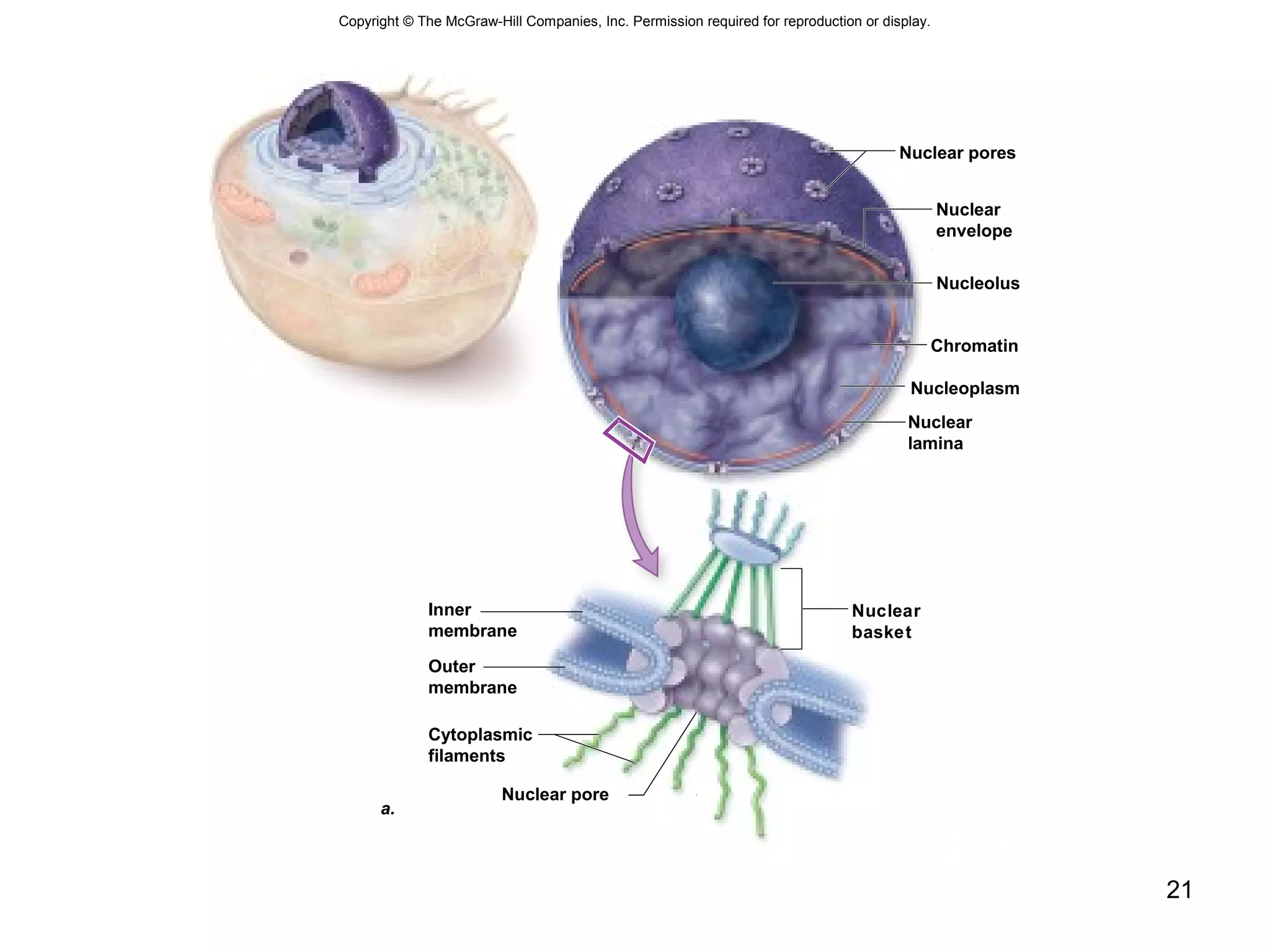
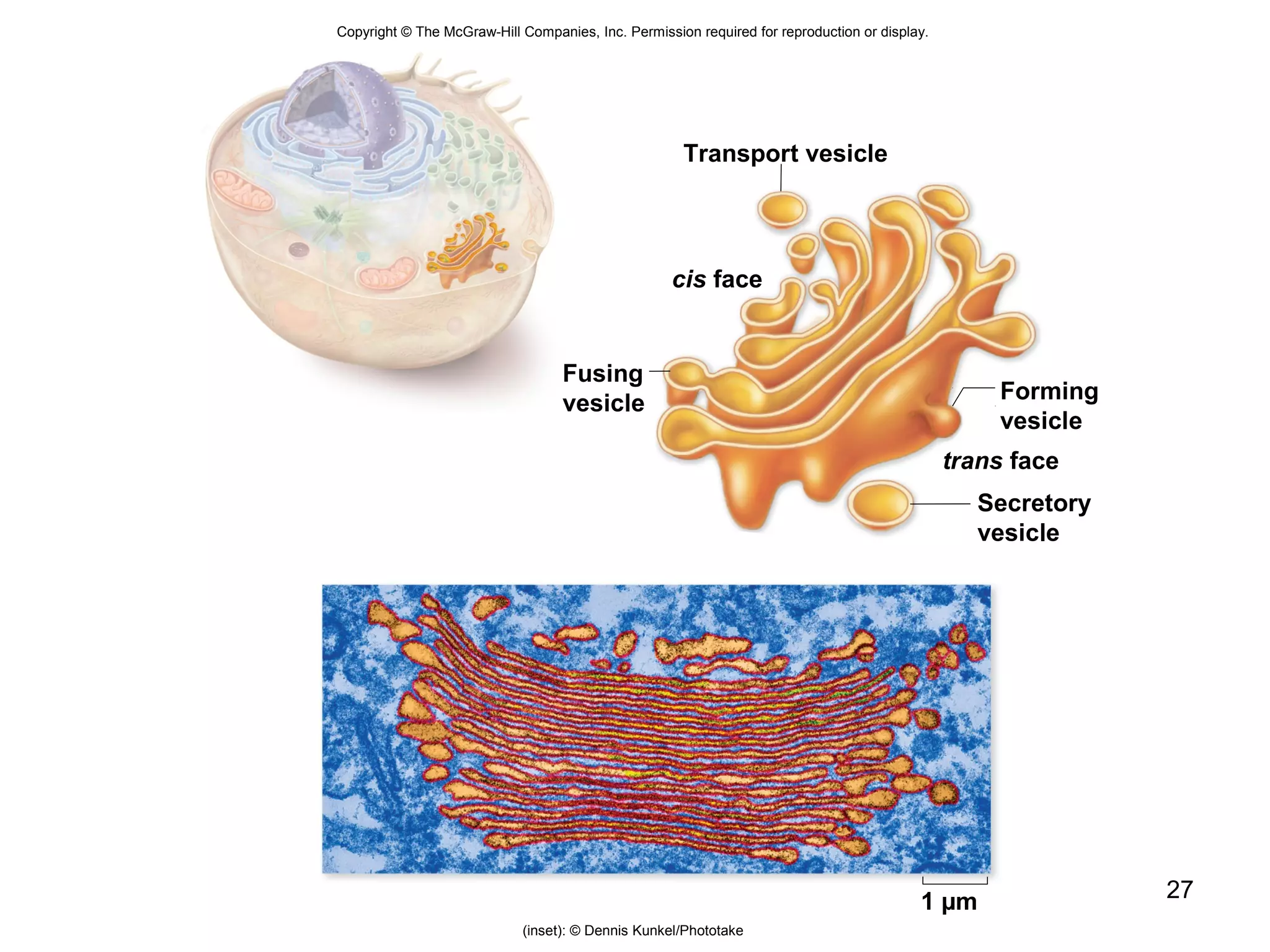
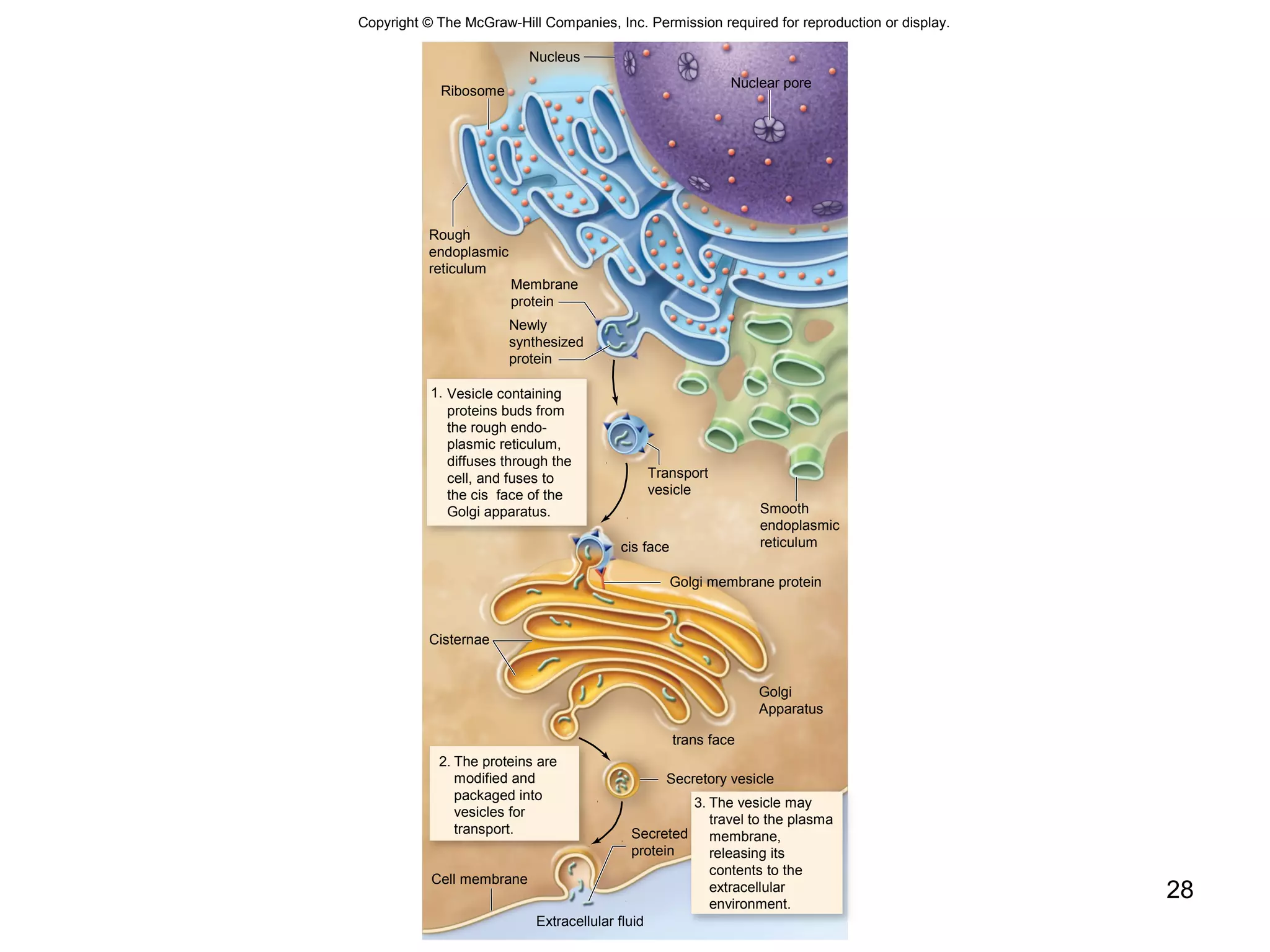
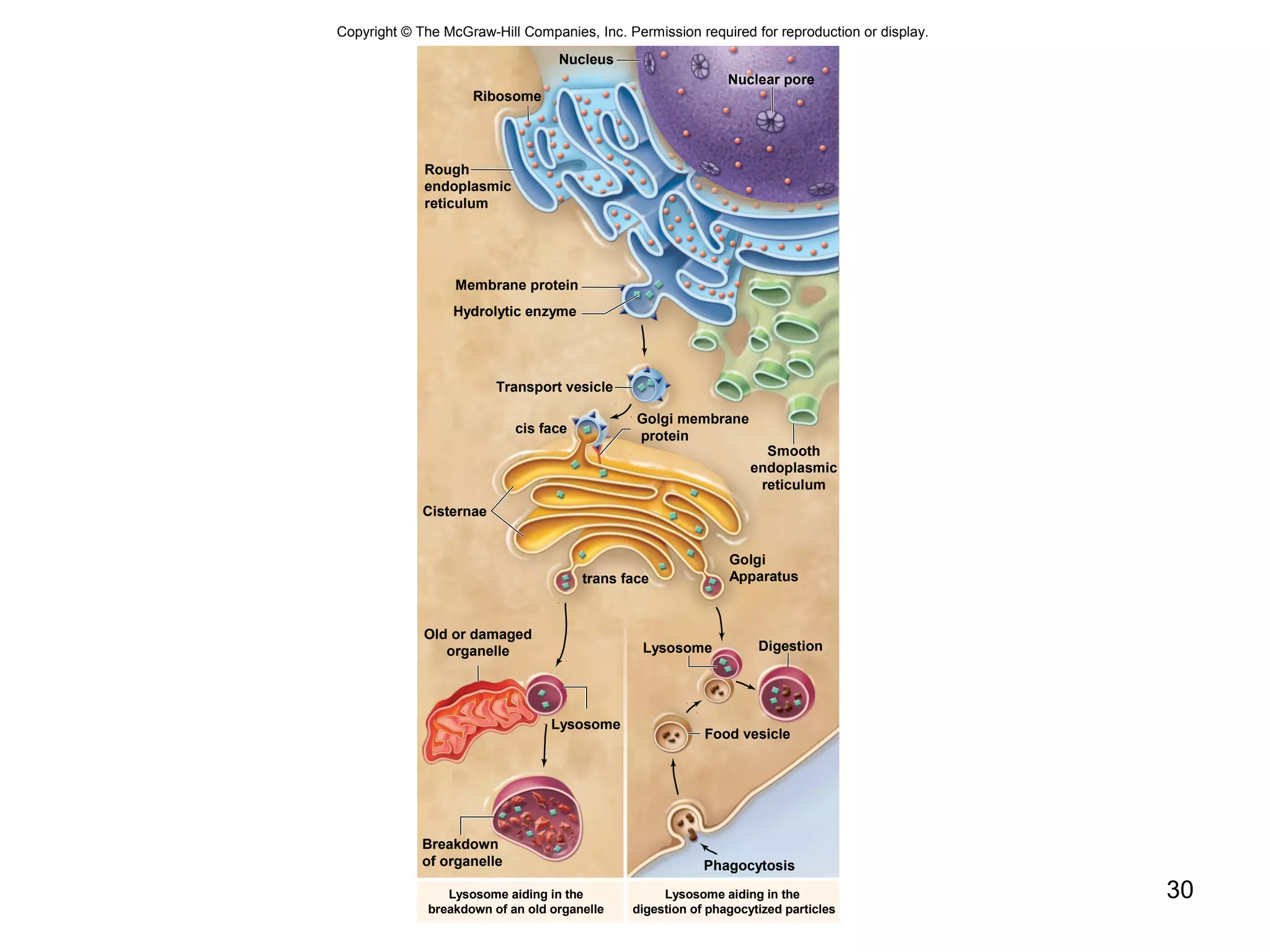
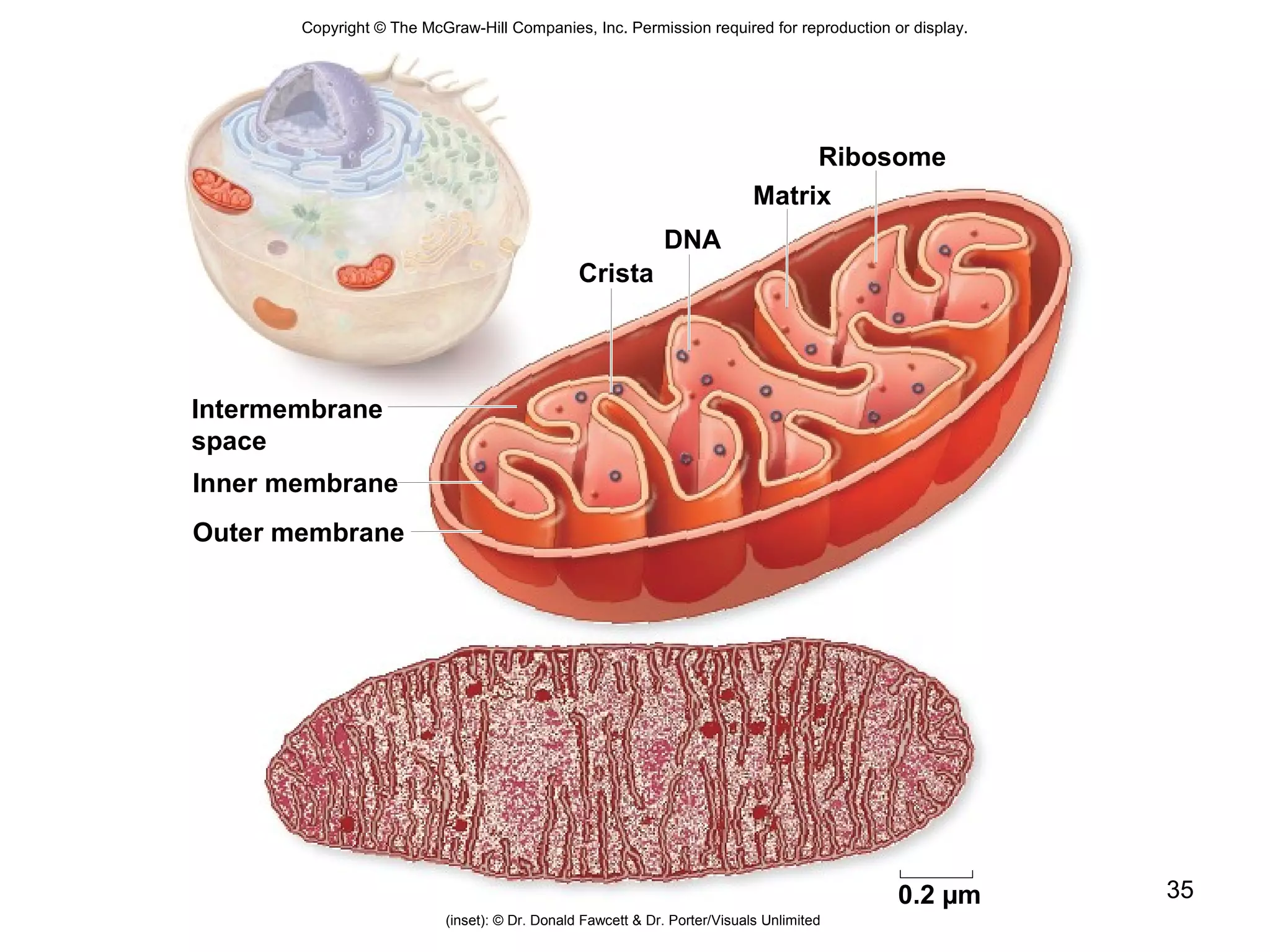
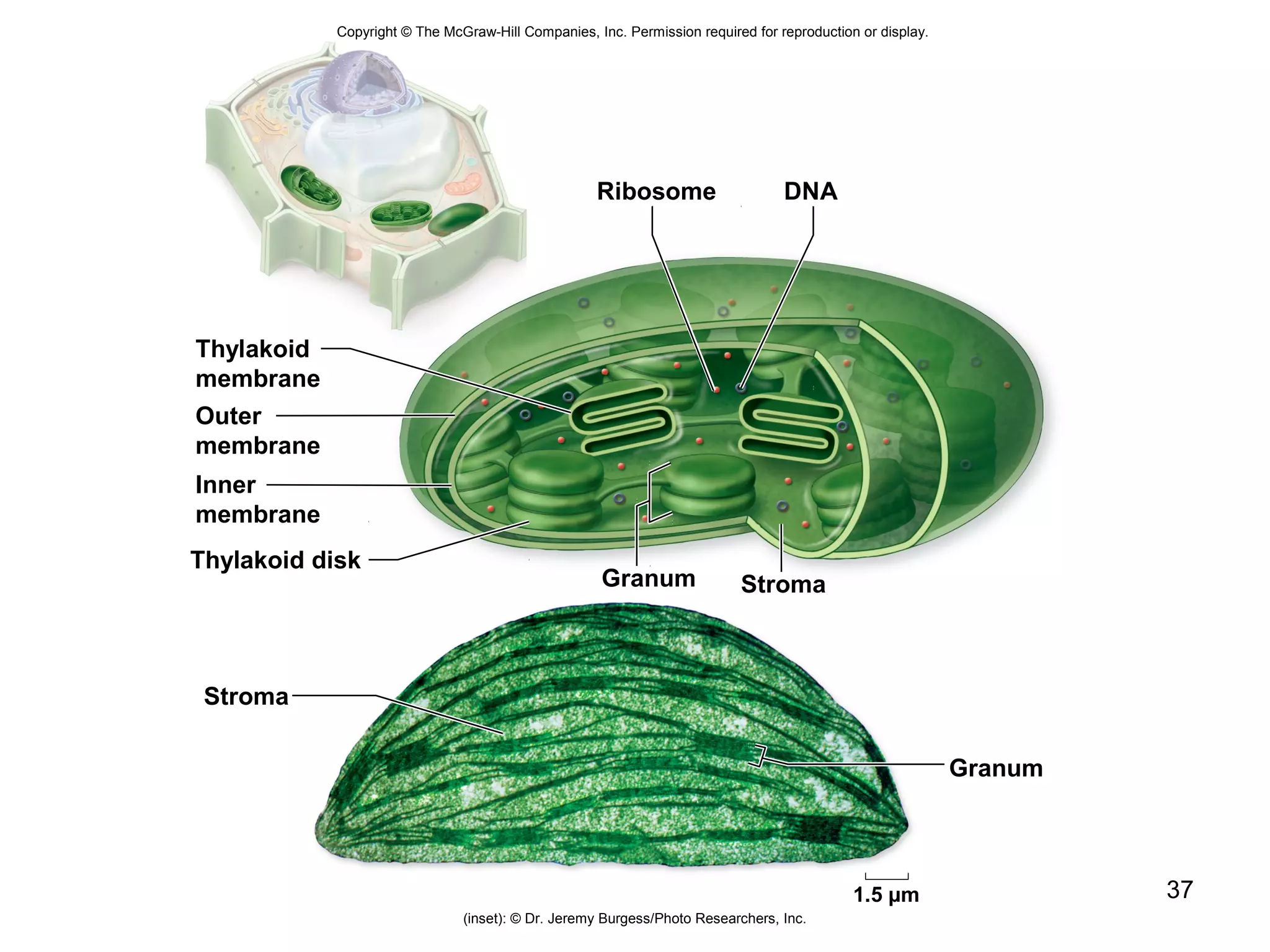
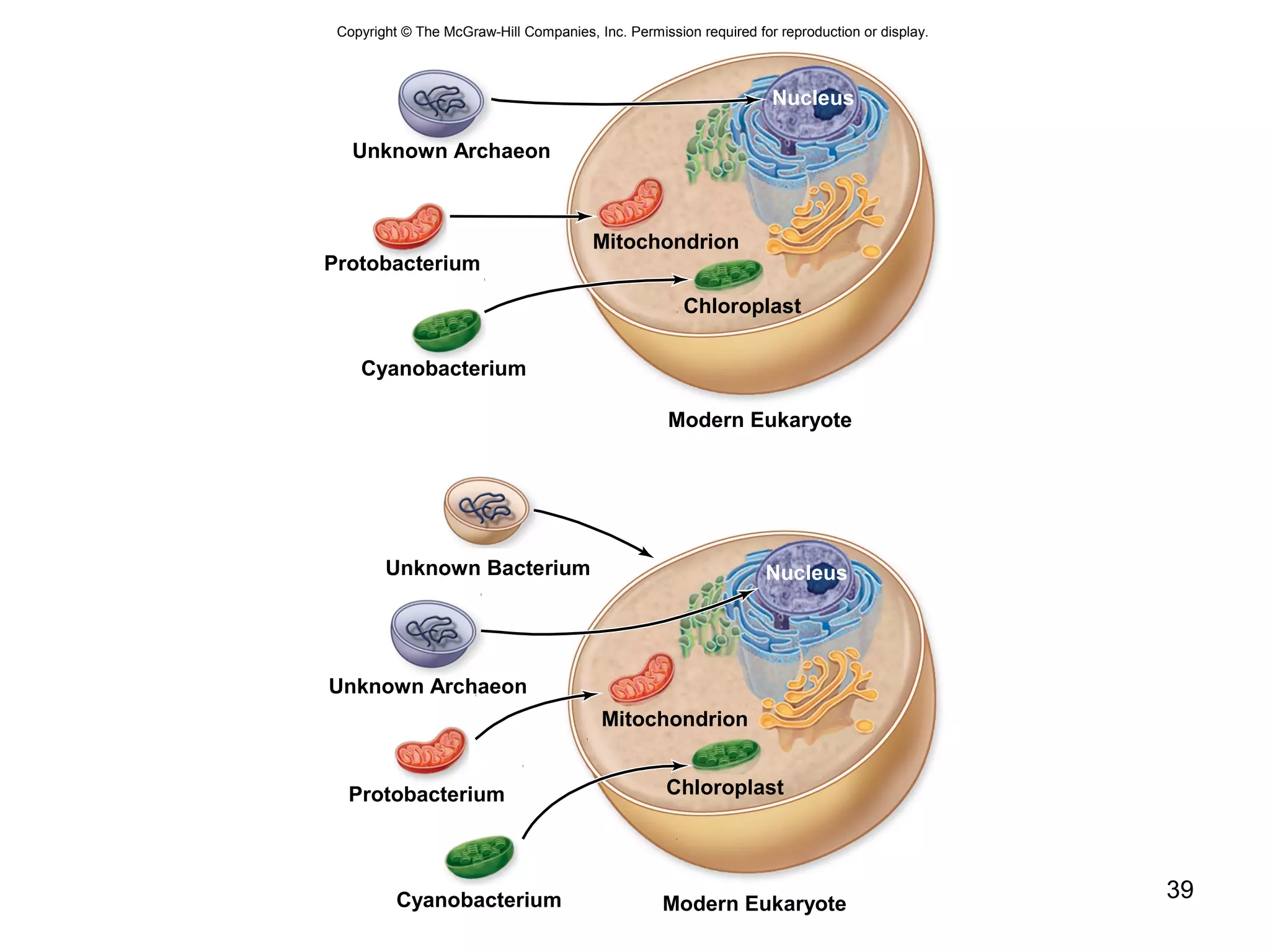
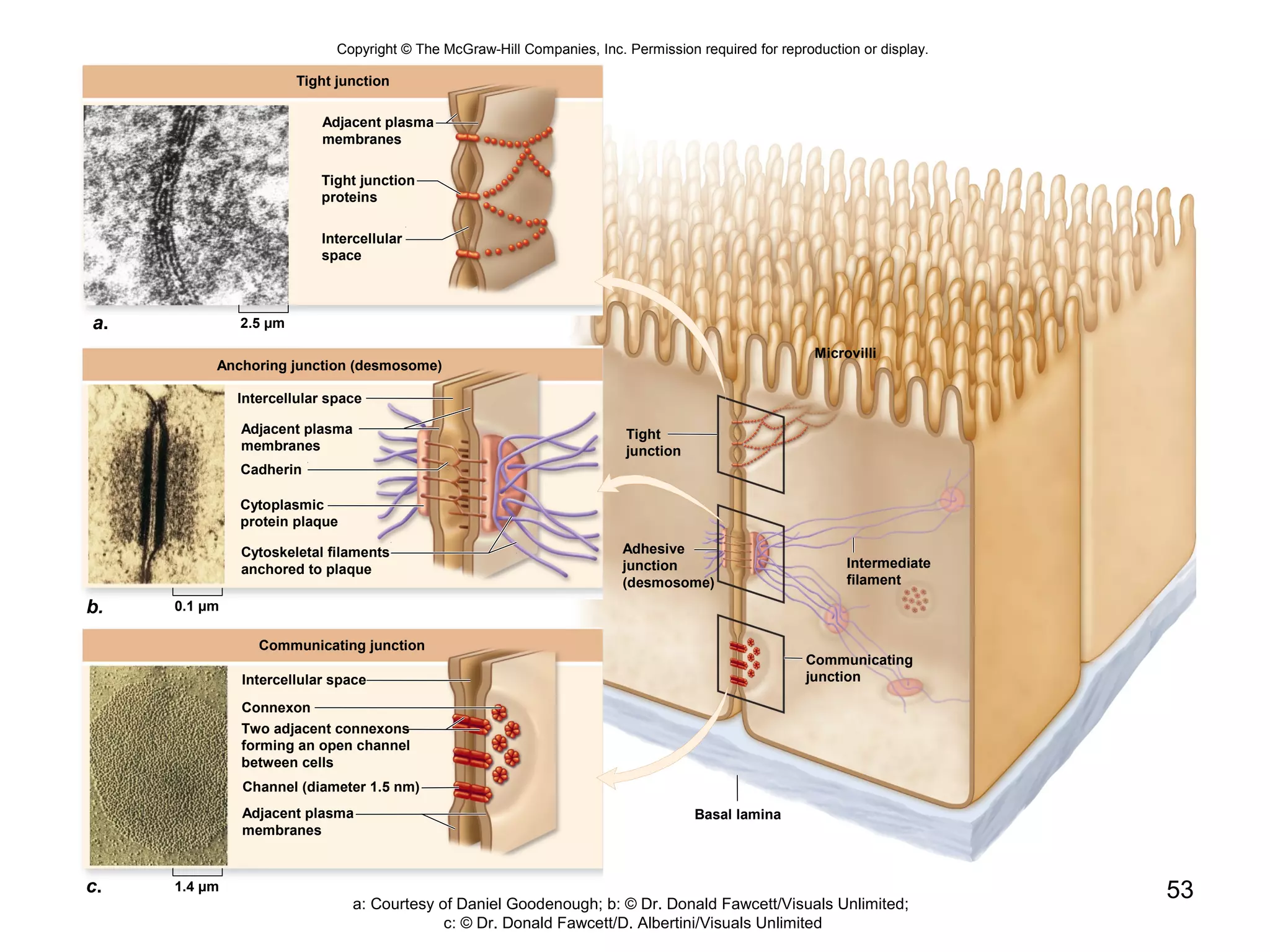
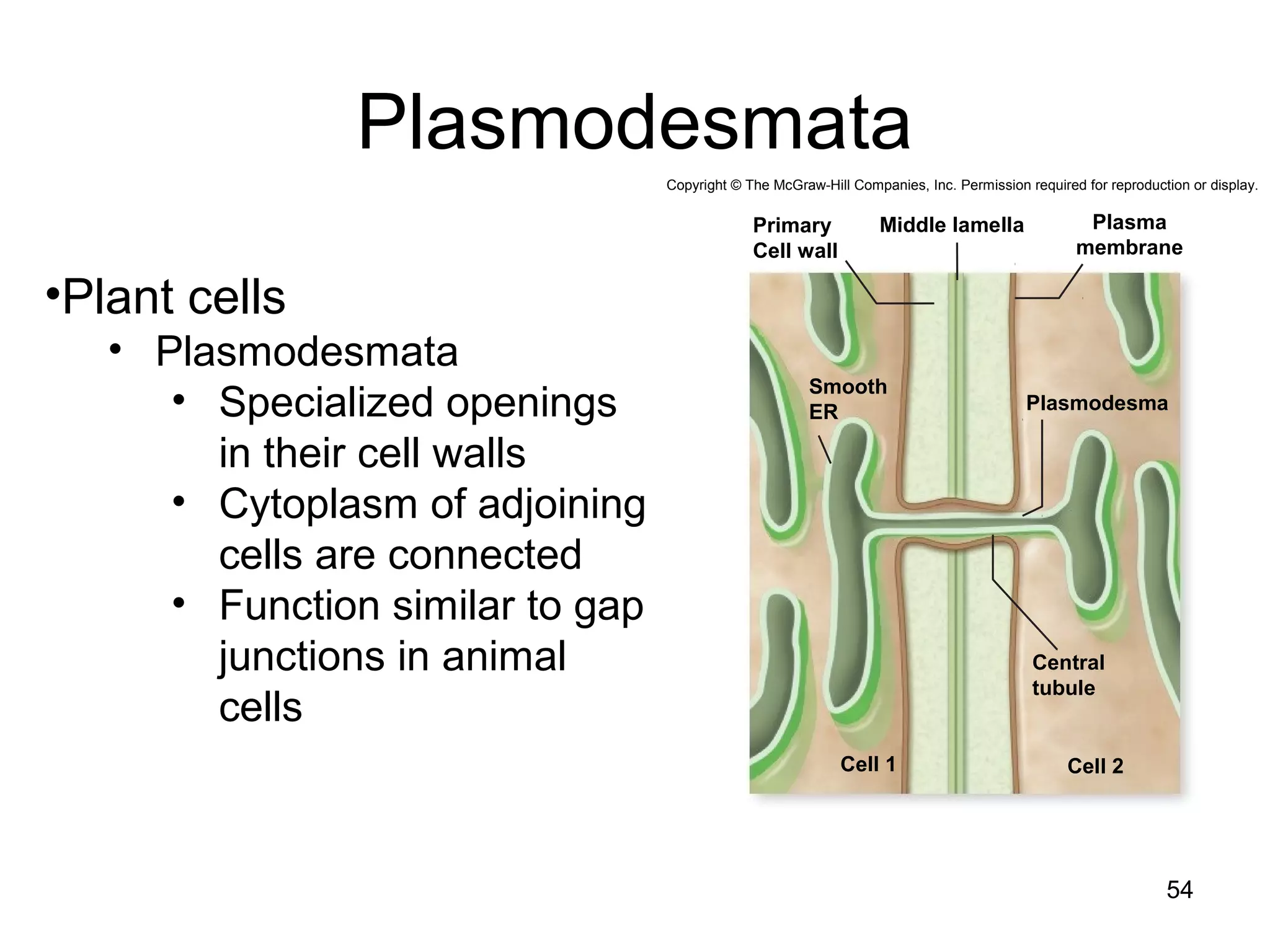
2. Both prokaryotic and eukaryotic cells have a plasma membrane, cytoplasm, and ribosomes. Eukaryotic cells also have organelles like the nucleus, mitochondria, chloroplasts, and Golgi apparatus.
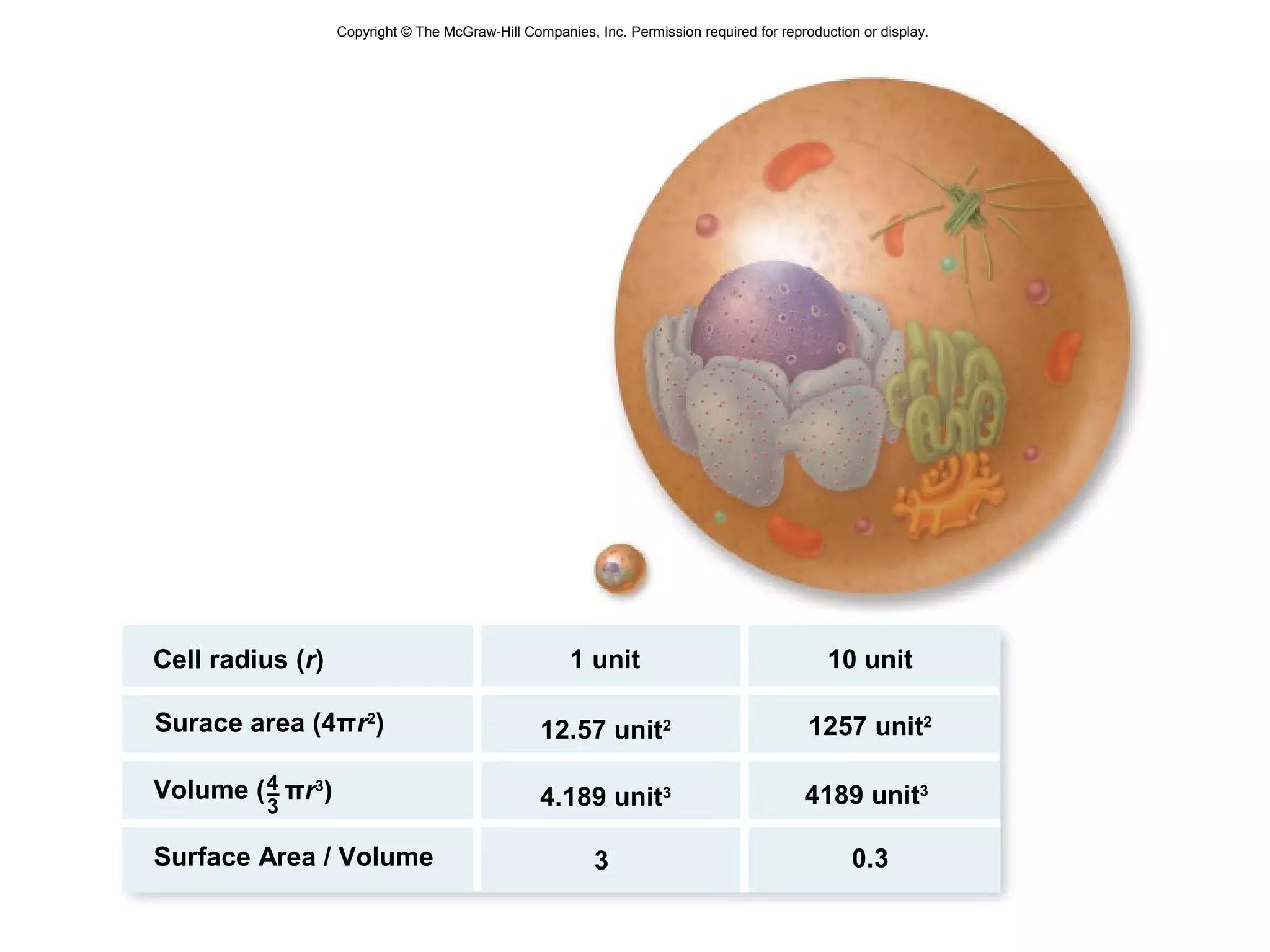
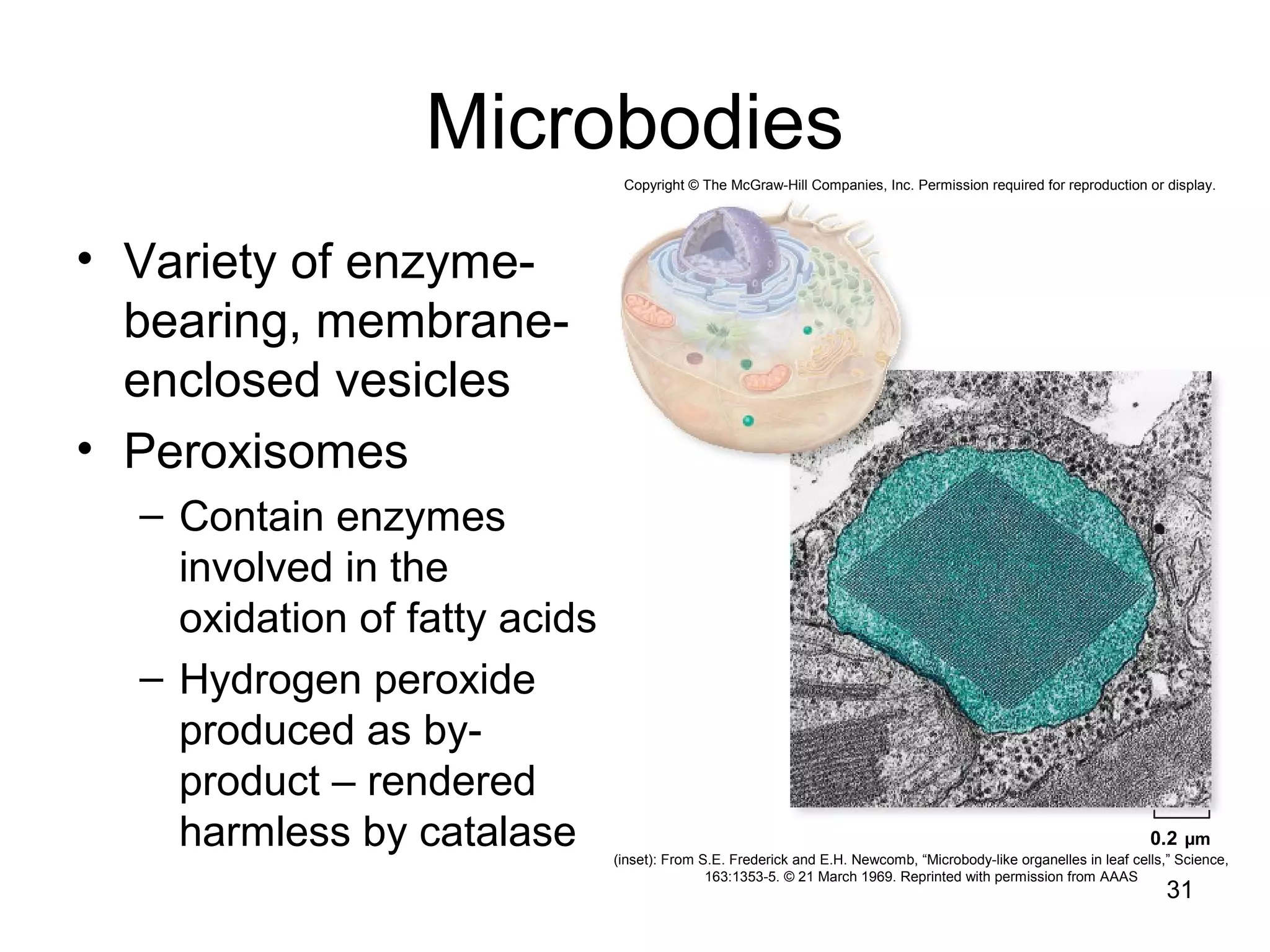
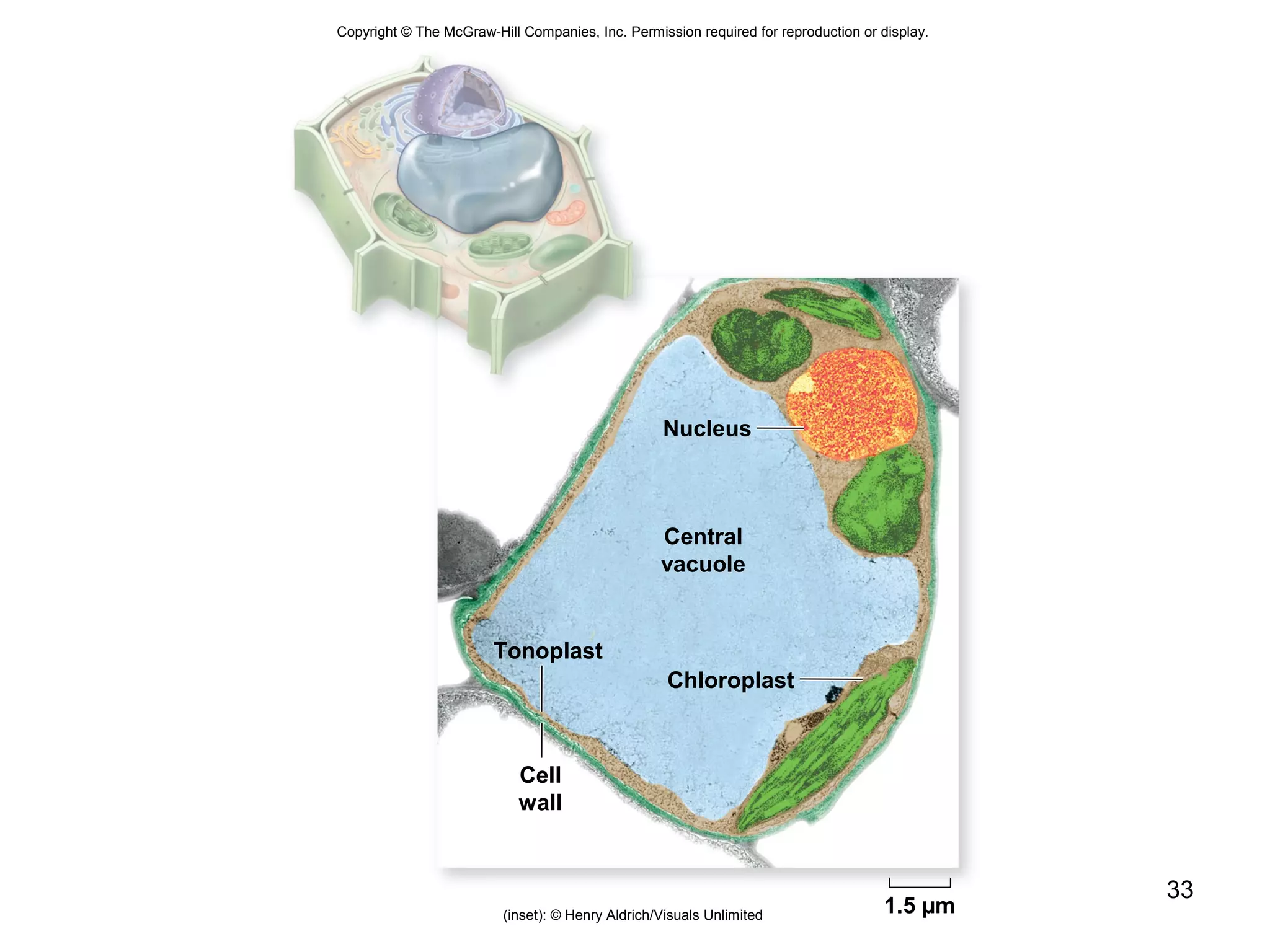
3. The size and structure of cells is limited by their need to exchange substances via diffusion. Organelles and compartments allow eukaryotic cells to perform specialized functions.