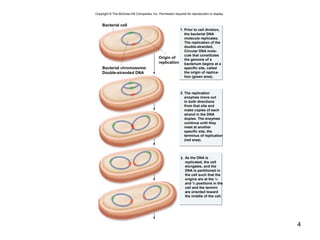
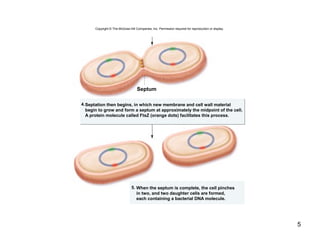
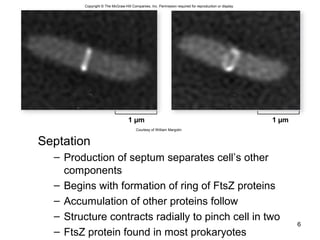
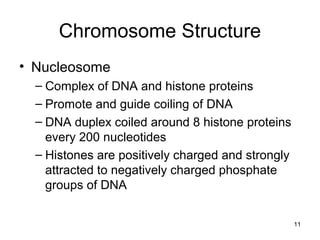
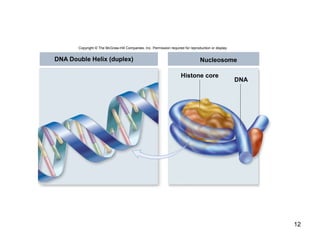
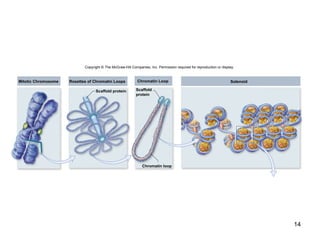
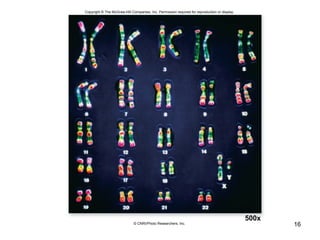
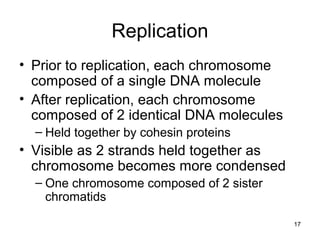
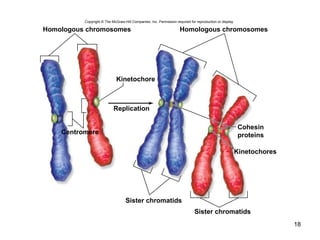
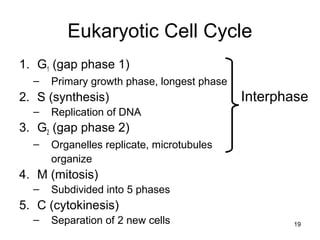
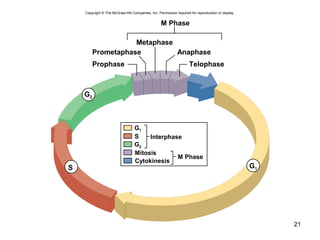
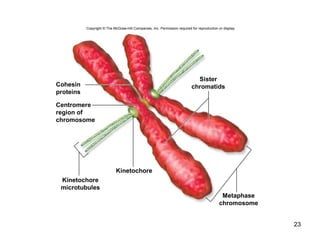
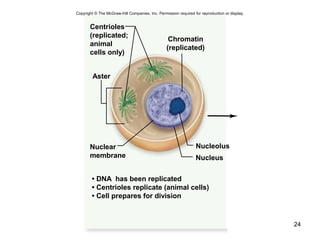
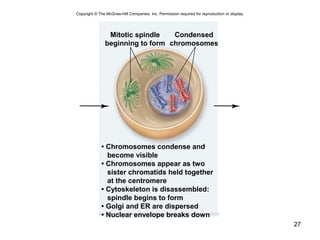
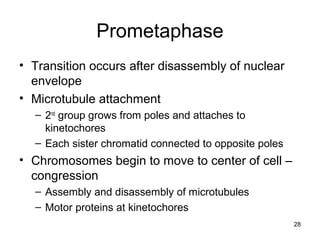
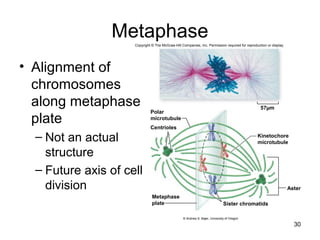
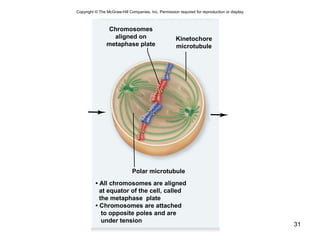
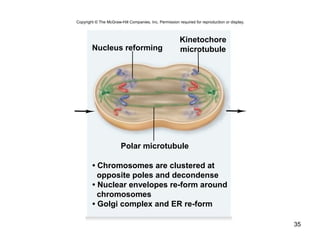
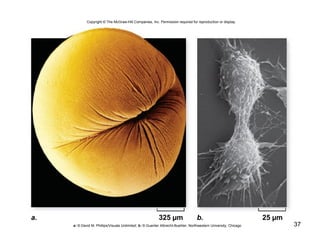
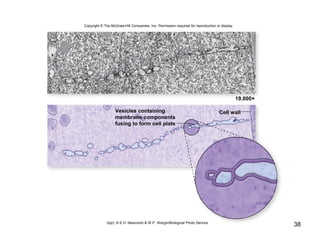
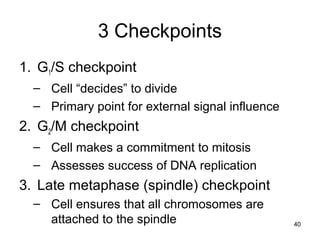
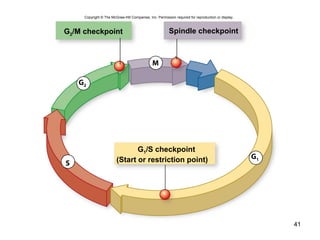
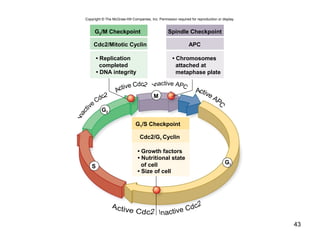
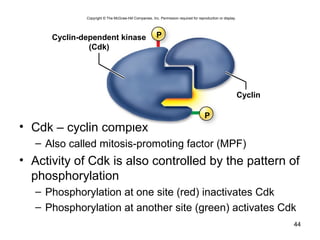
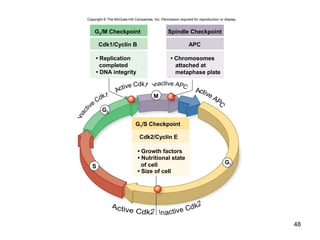
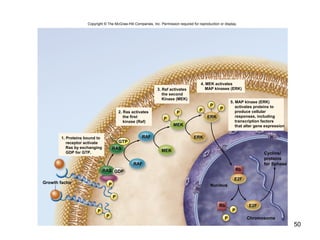
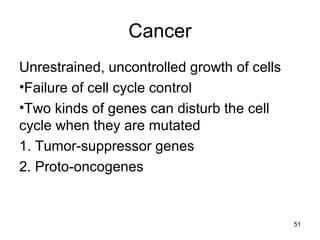
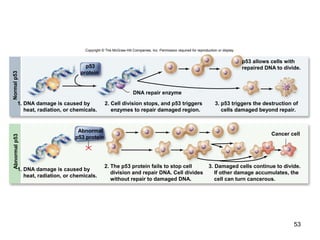
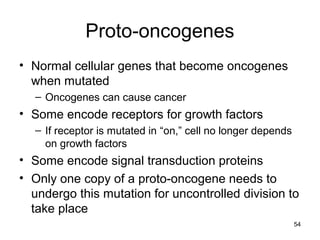
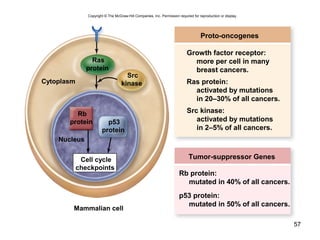
This document provides an outline and overview of chapter 10 on cell division. It discusses bacterial cell division through binary fission and the replication of the bacterial chromosome. It then summarizes eukaryotic cell division, including the stages of mitosis (prophase, prometaphase, metaphase, anaphase, telophase) and cytokinesis. It also discusses the structure and compaction of eukaryotic chromosomes, the key events of the cell cycle including checkpoints, and the role of cyclin-dependent kinases in regulating the cell cycle. The document is accompanied by figures and tables to illustrate the concepts discussed.