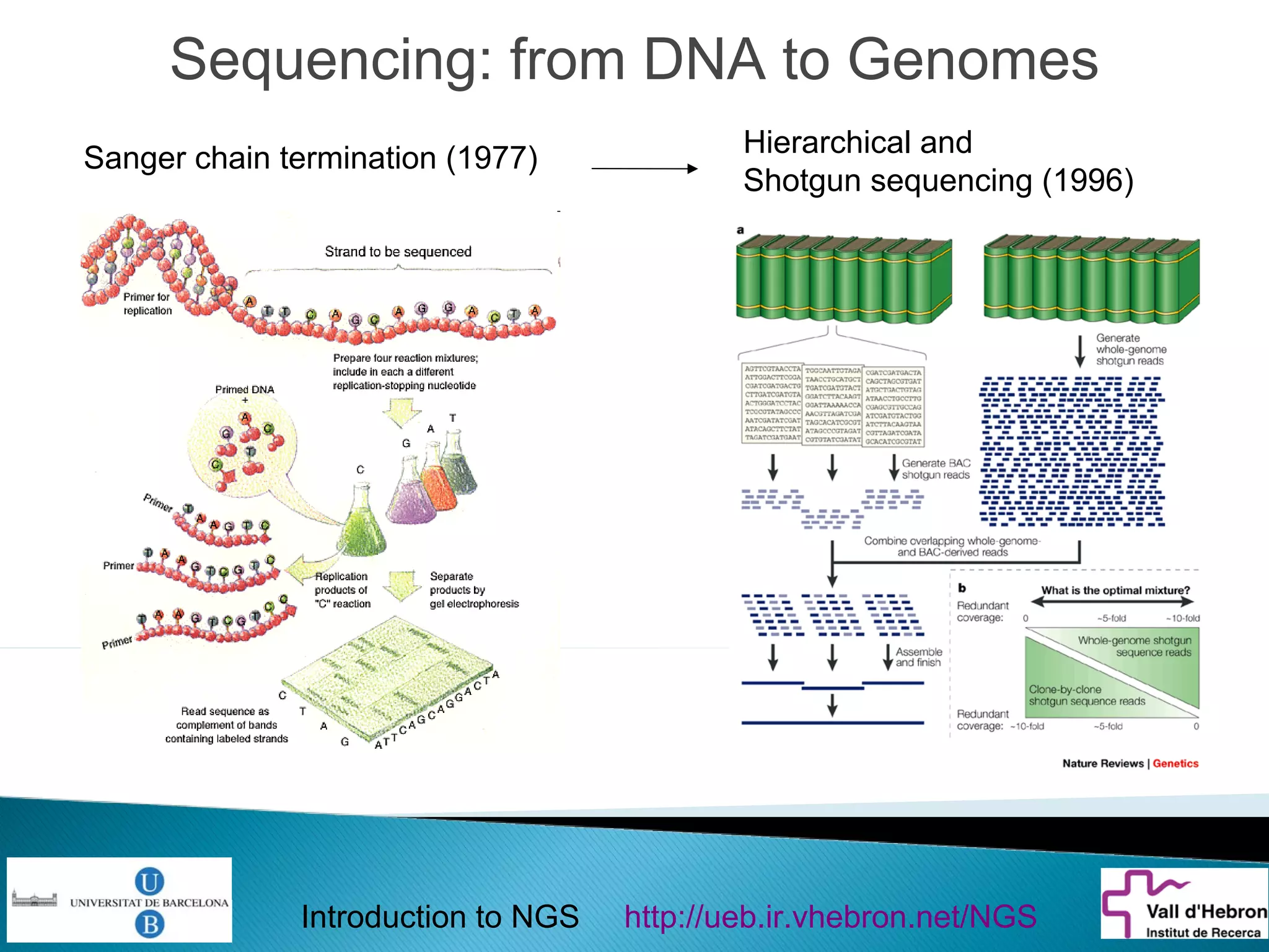
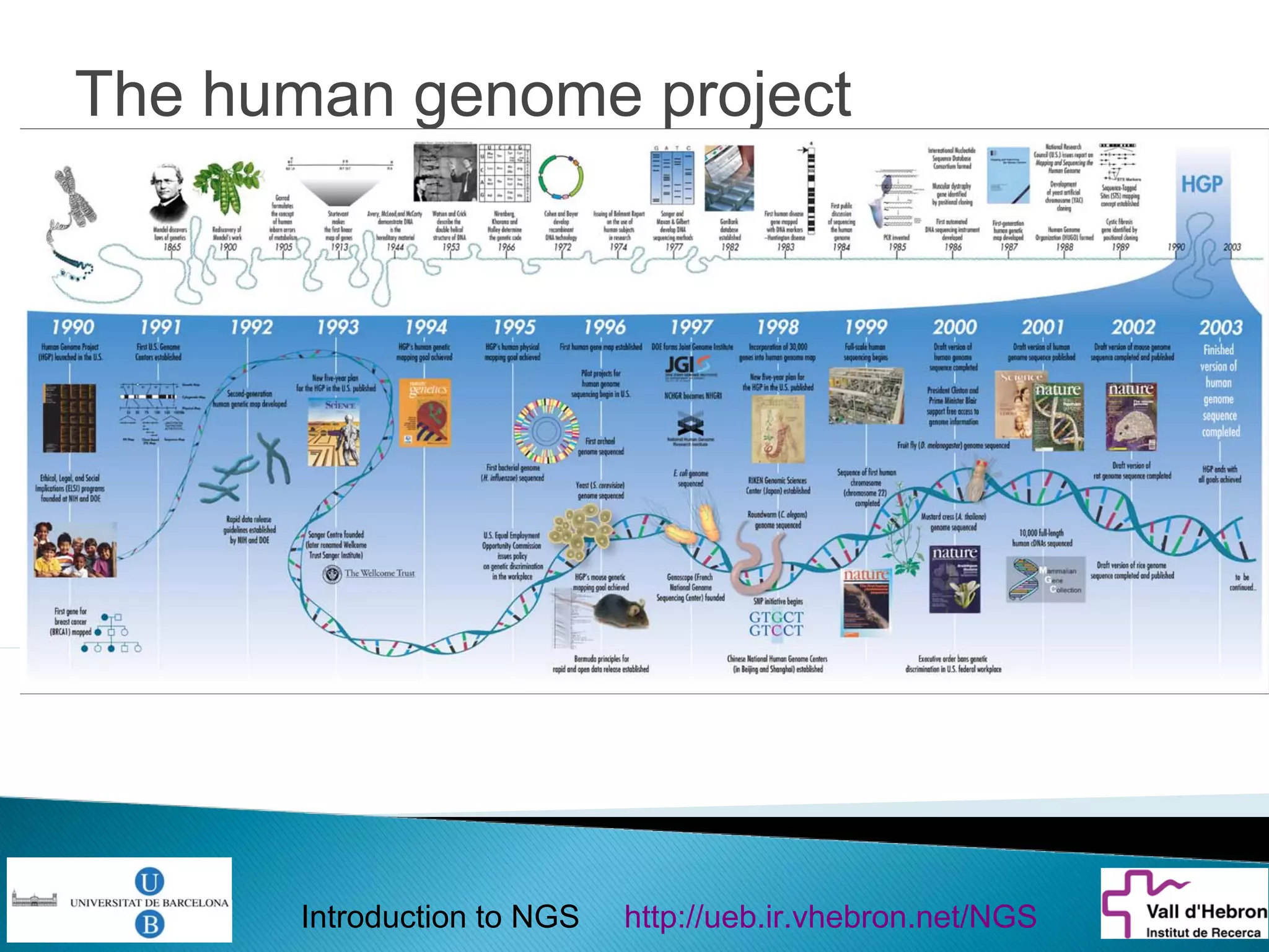
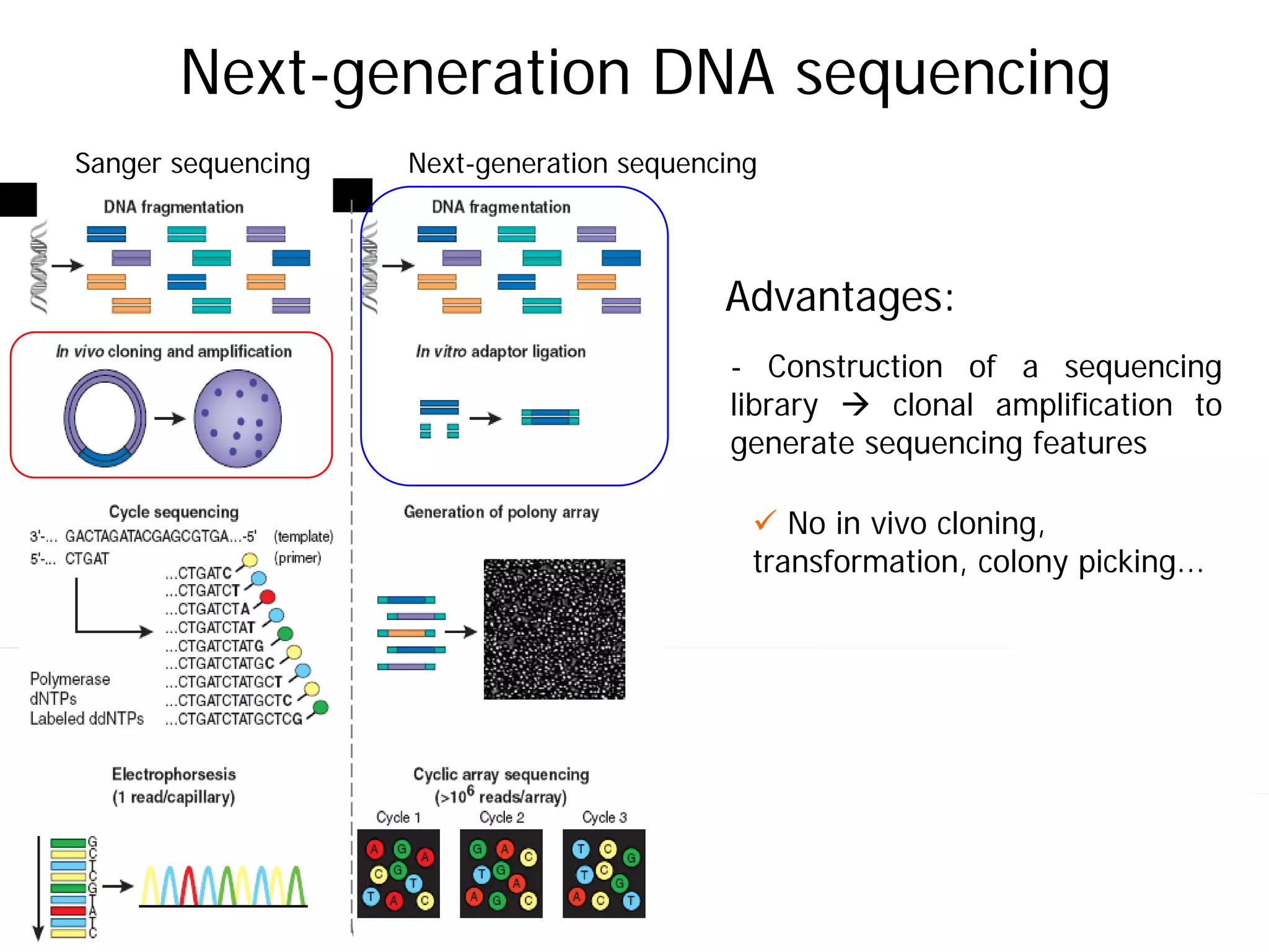
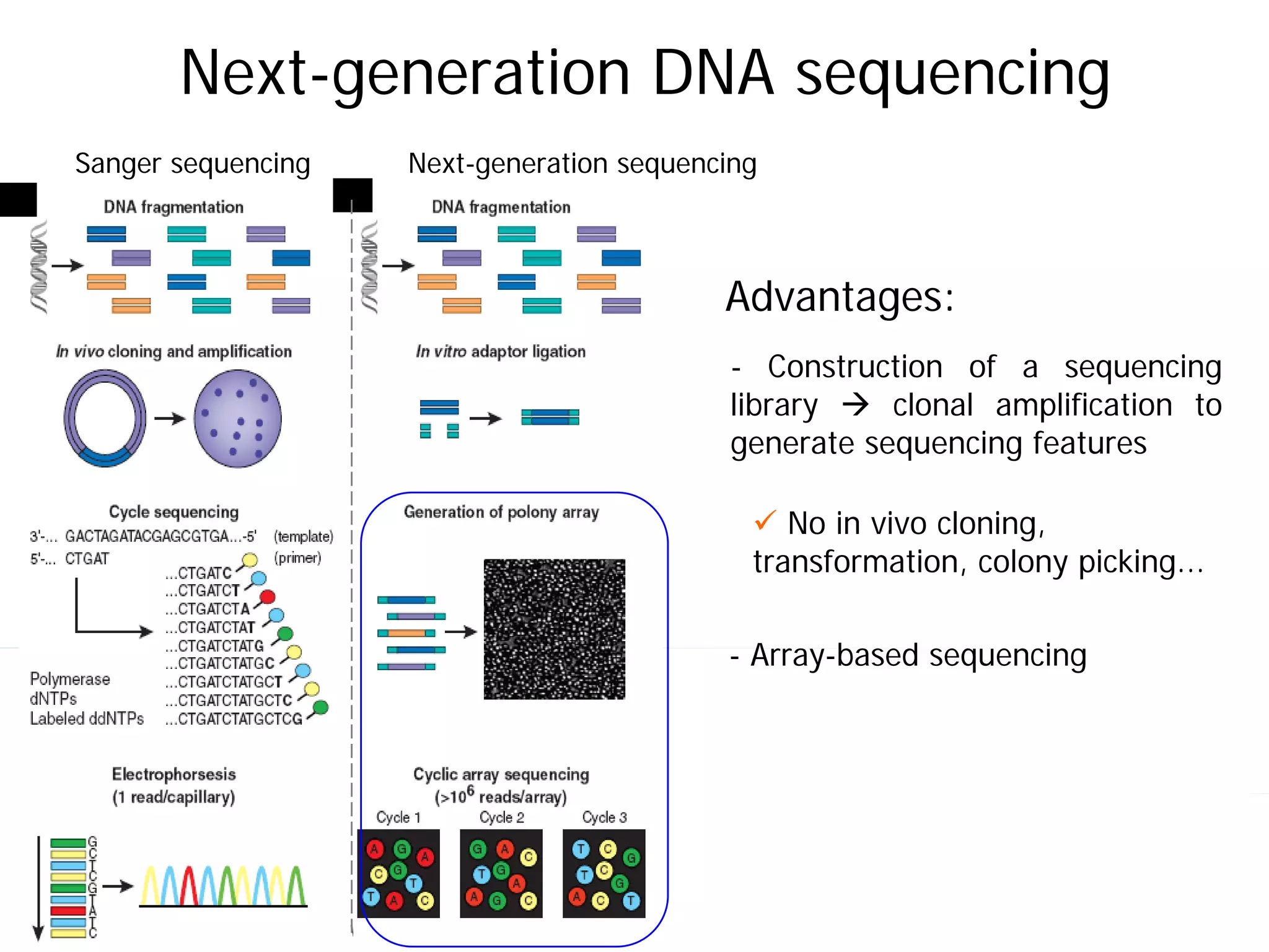
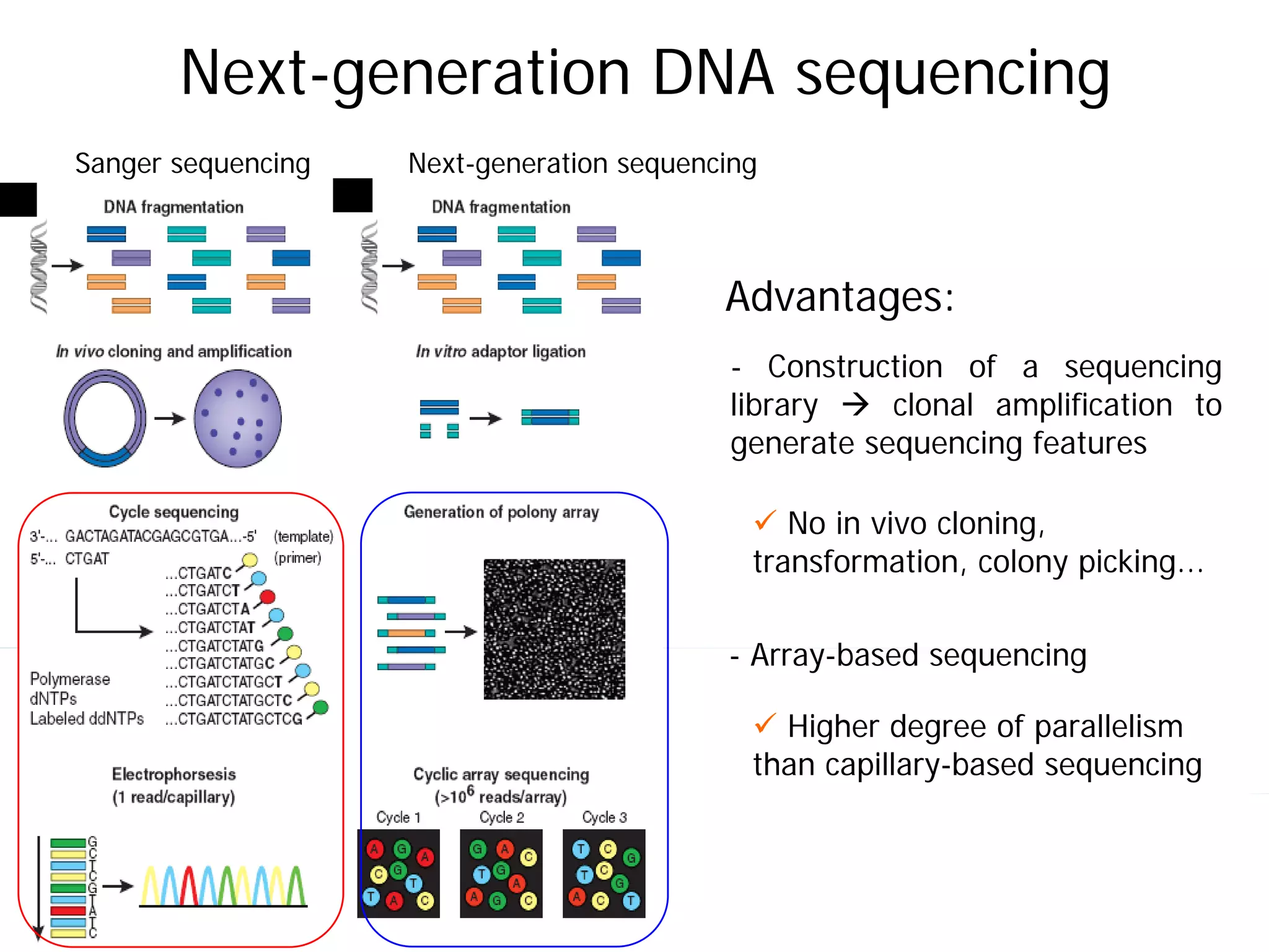
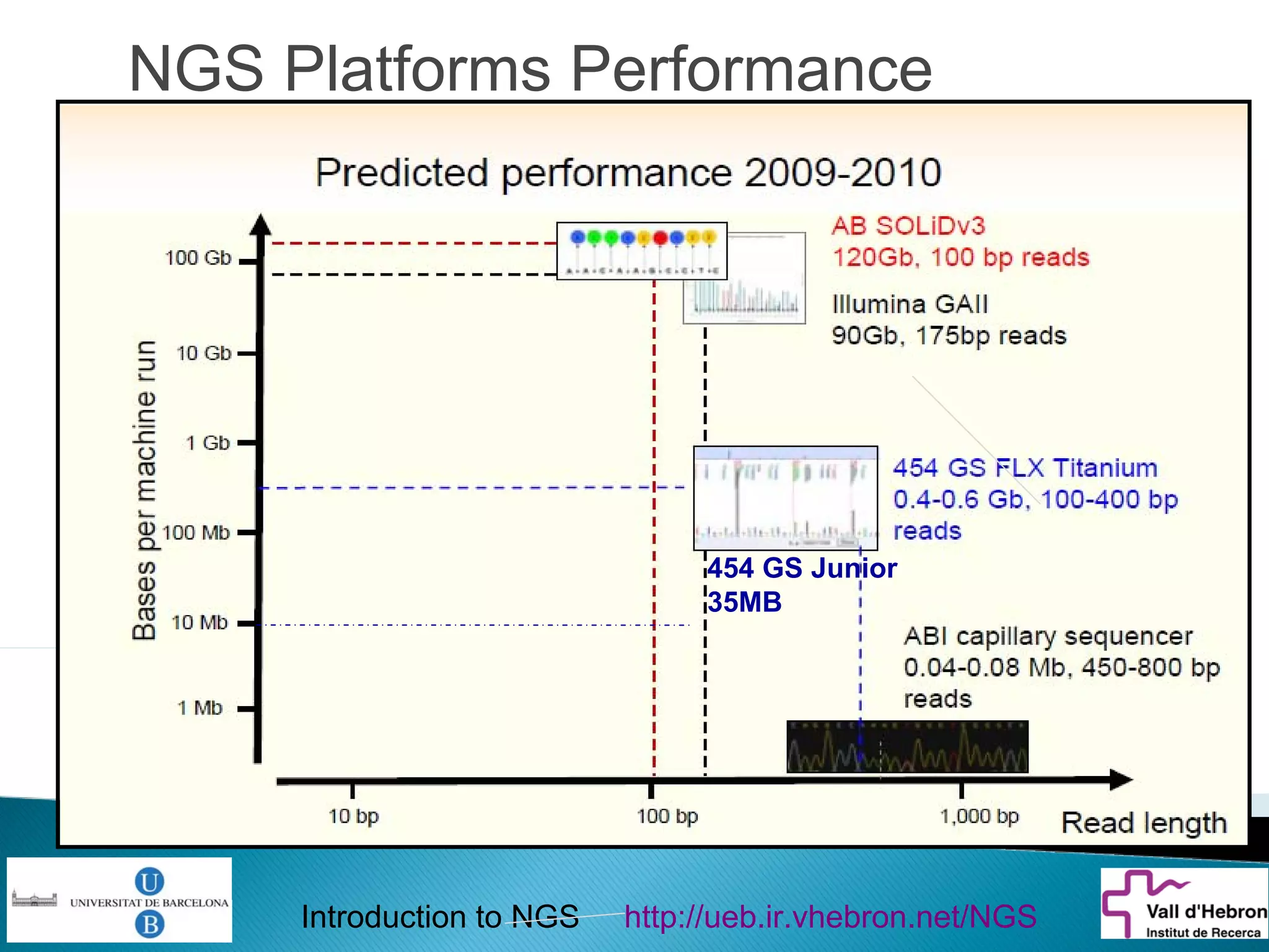
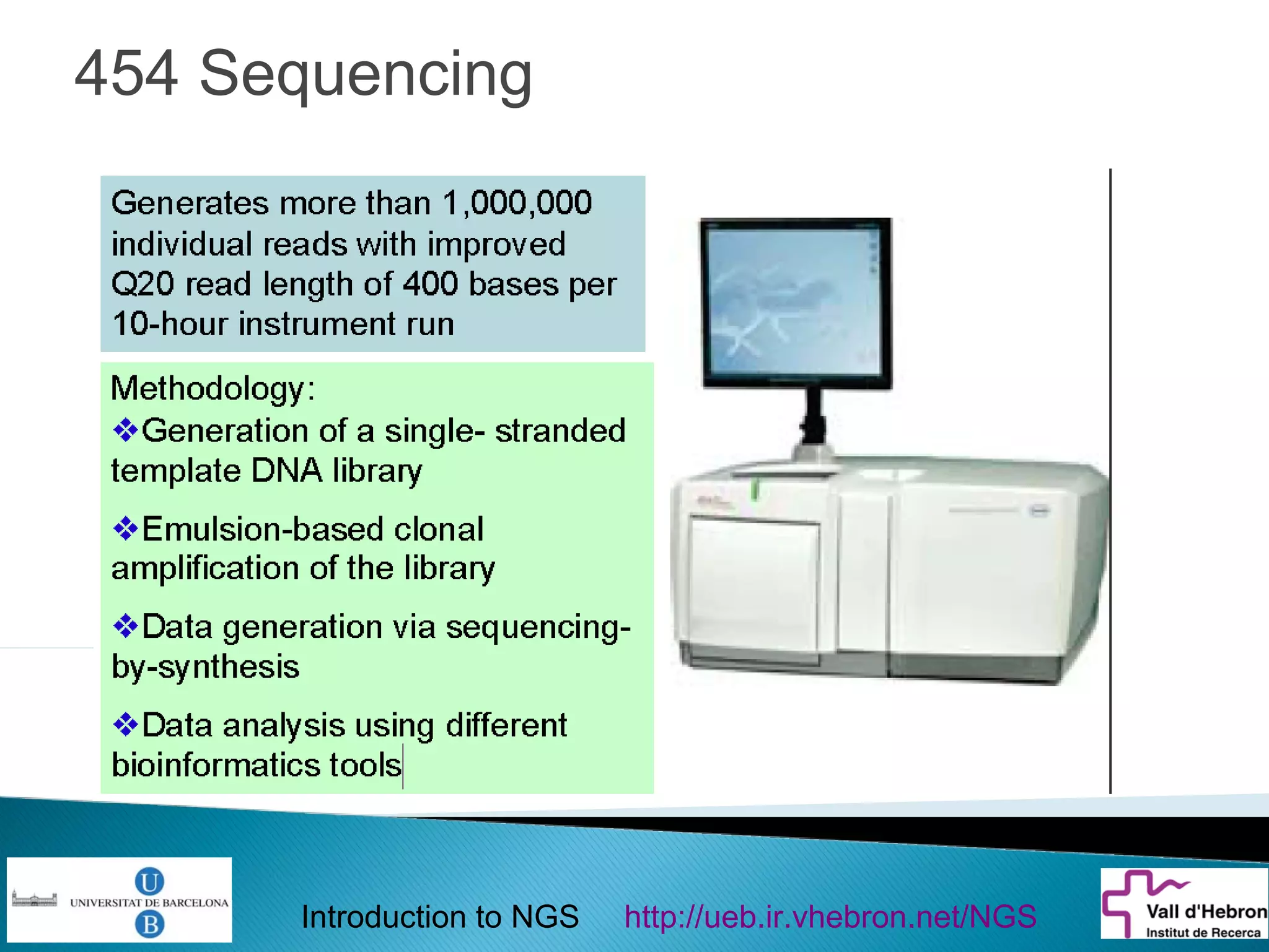
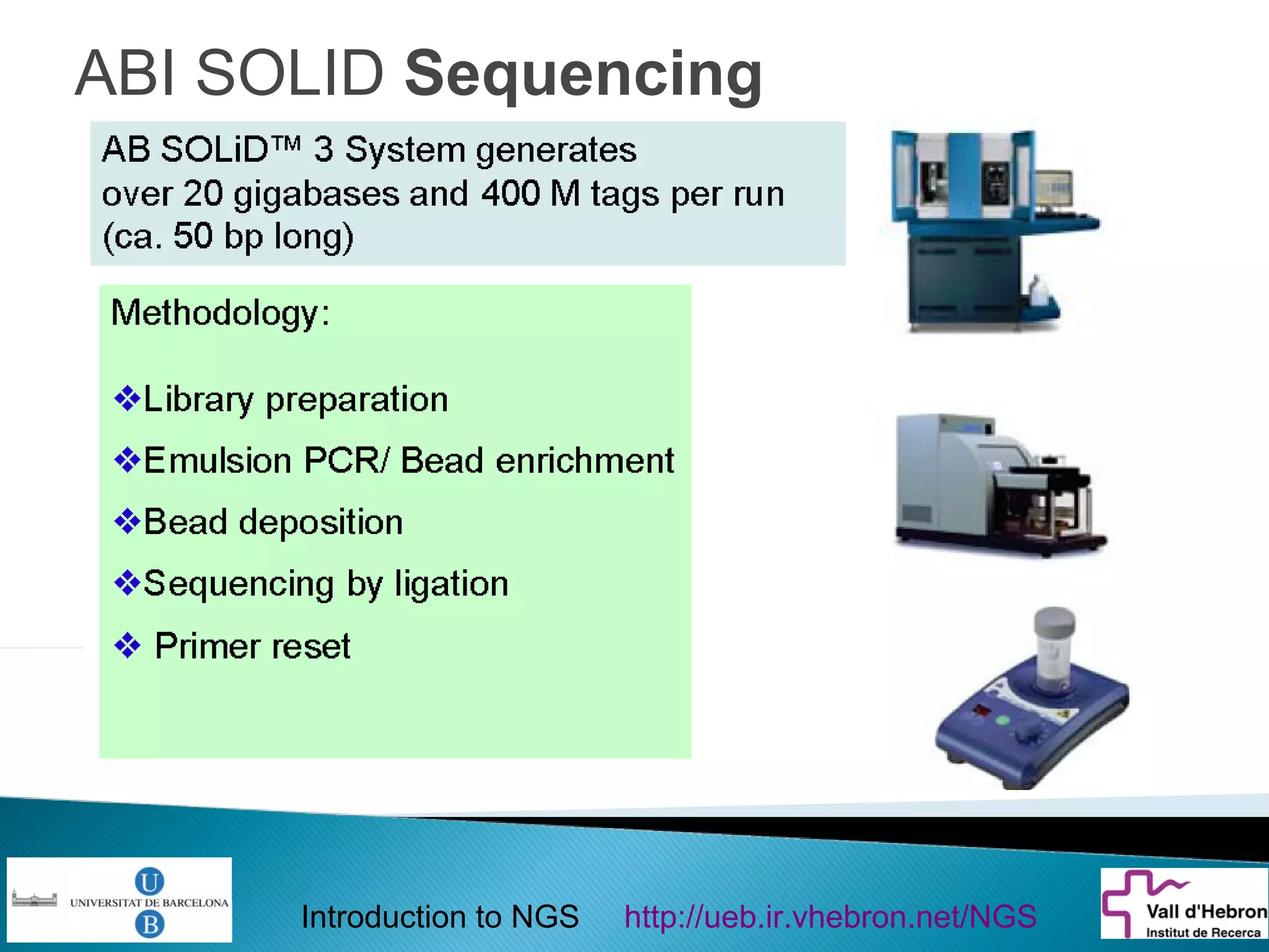
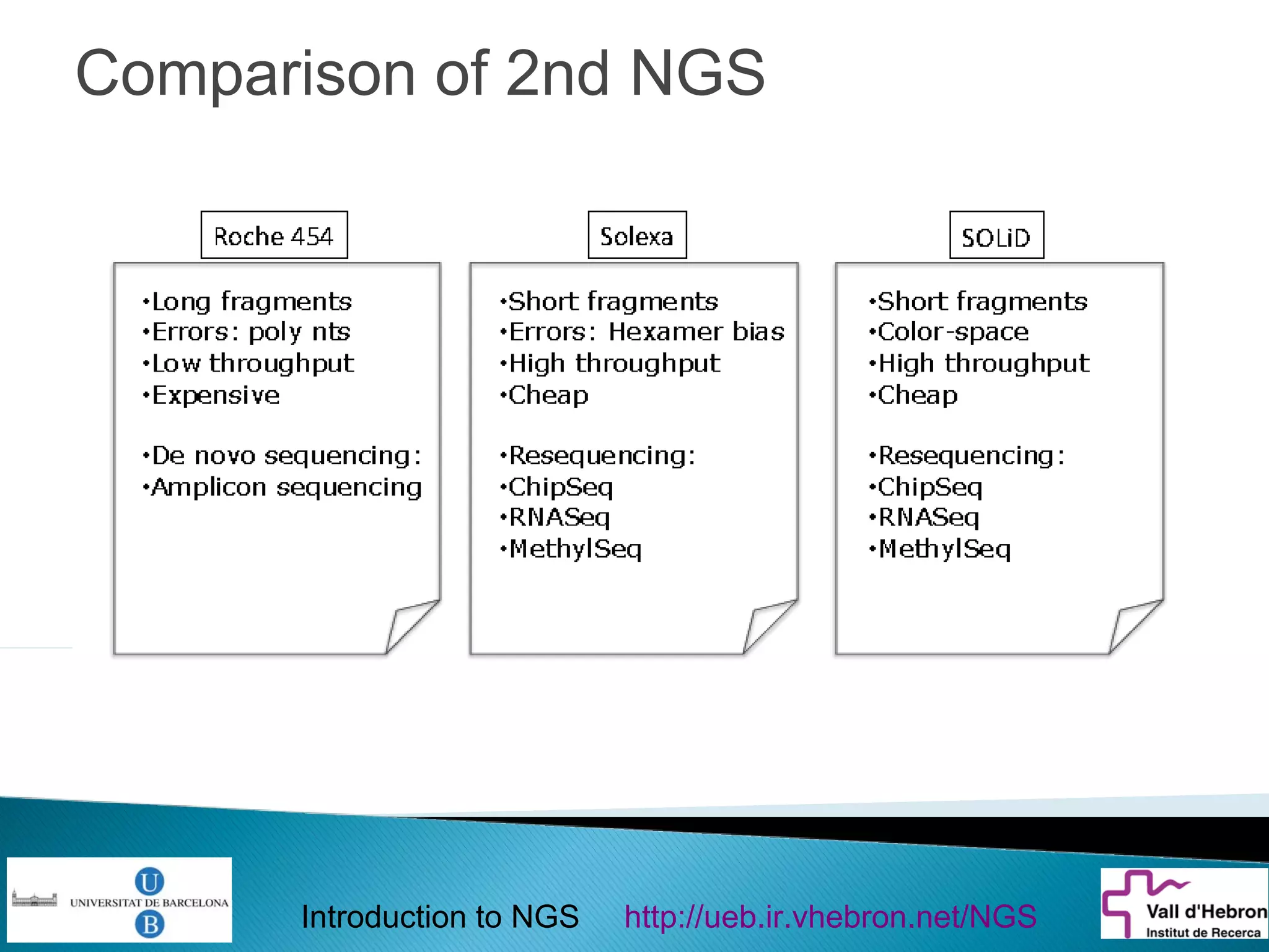
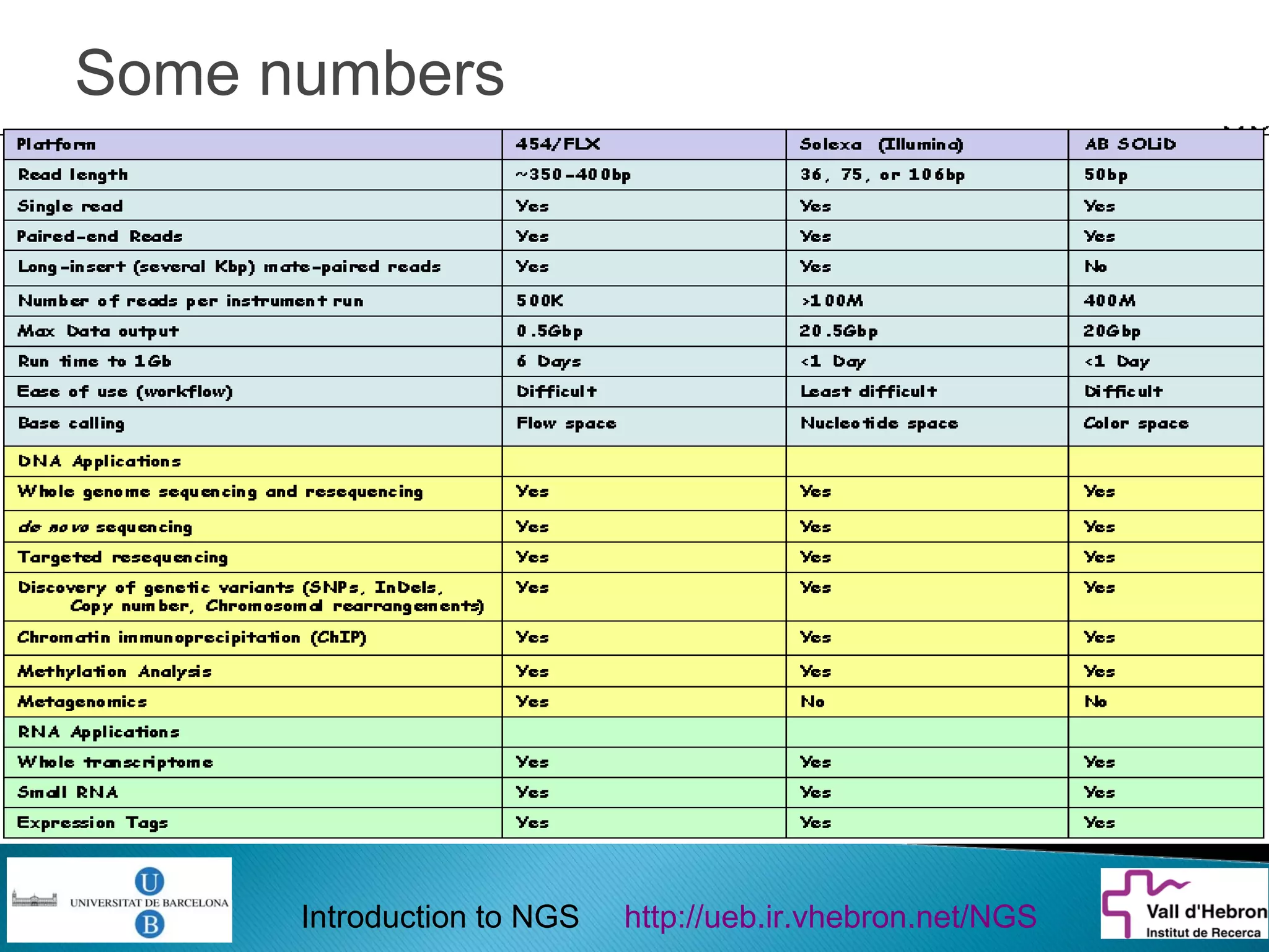
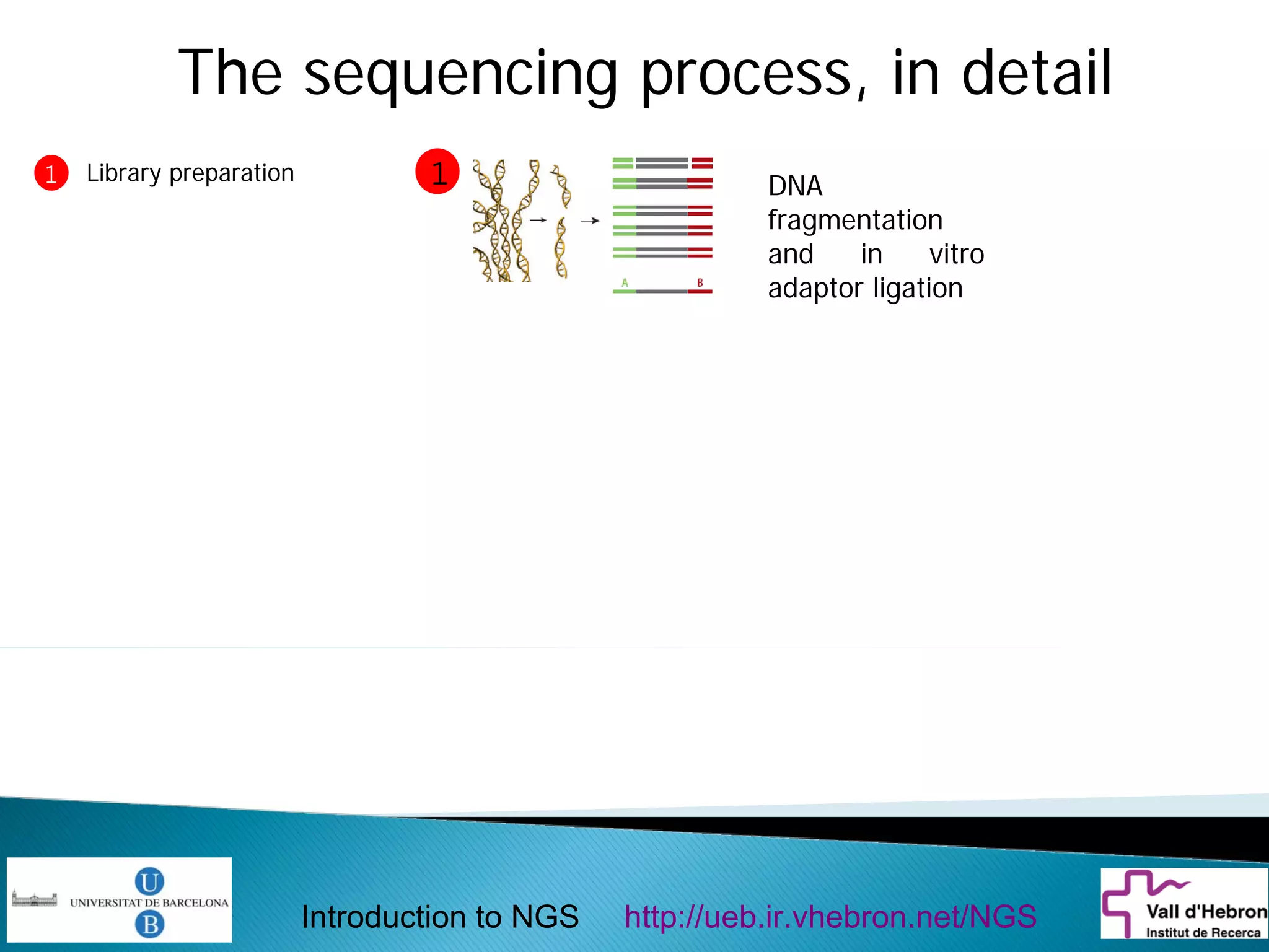
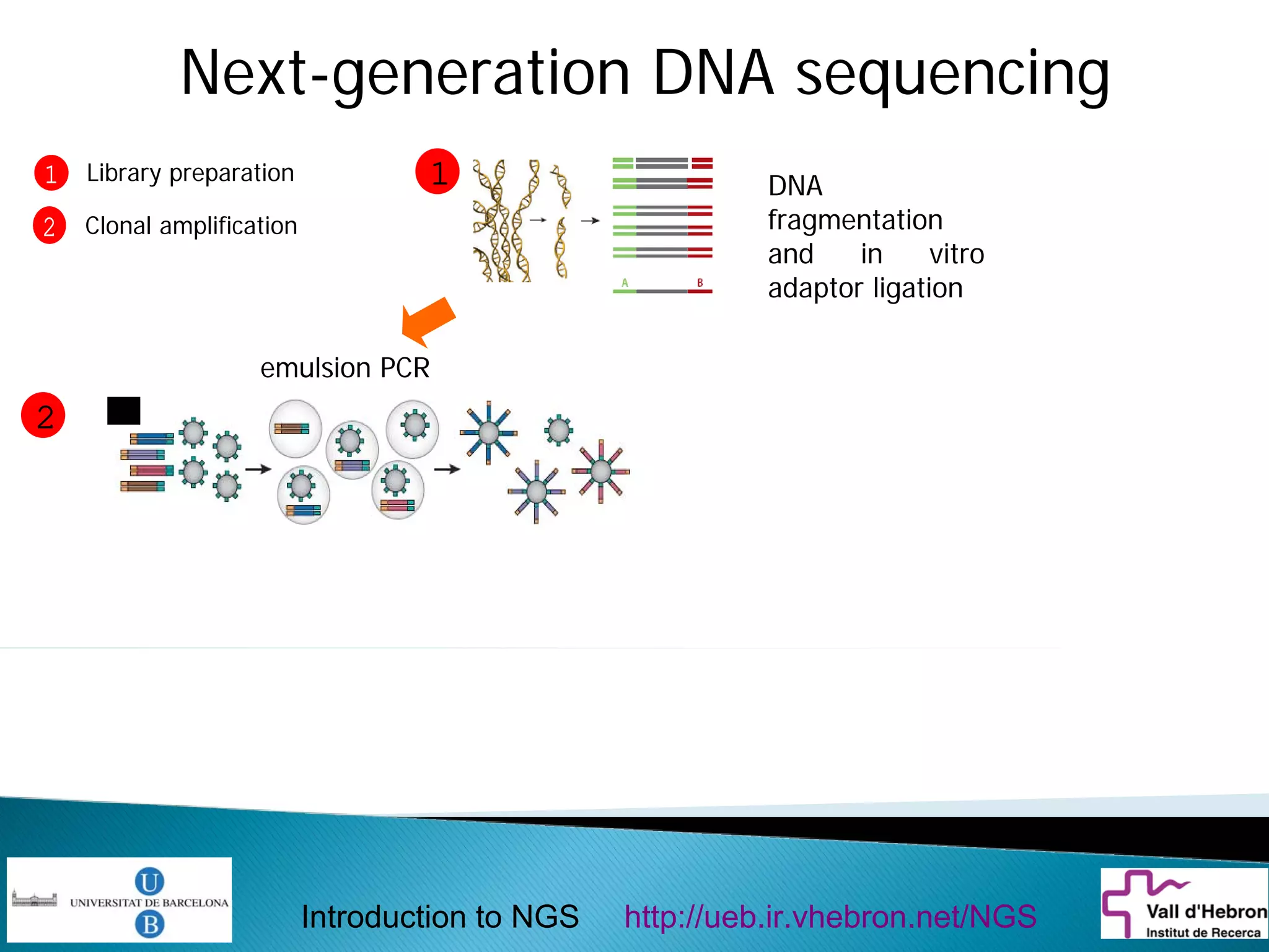
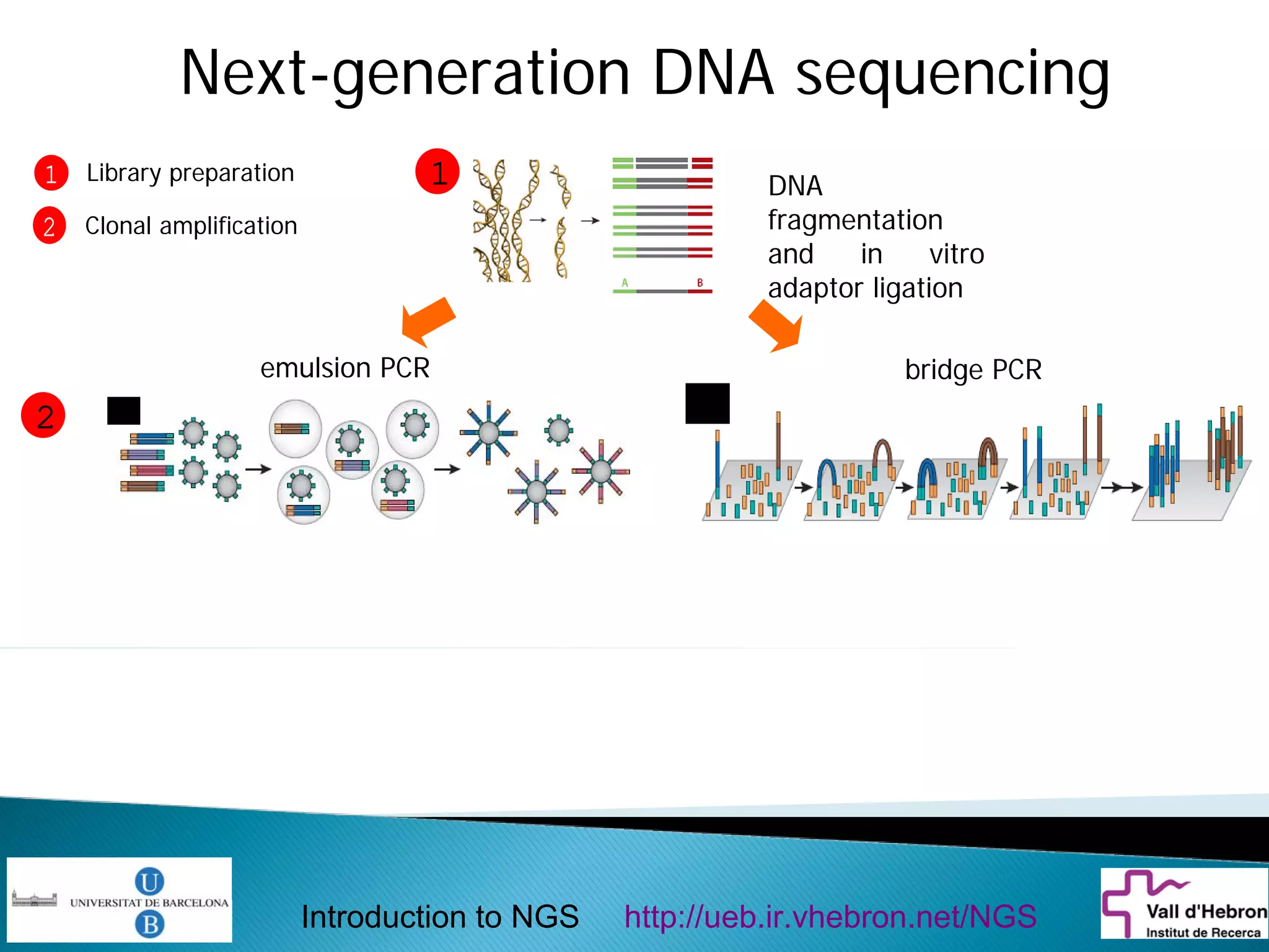
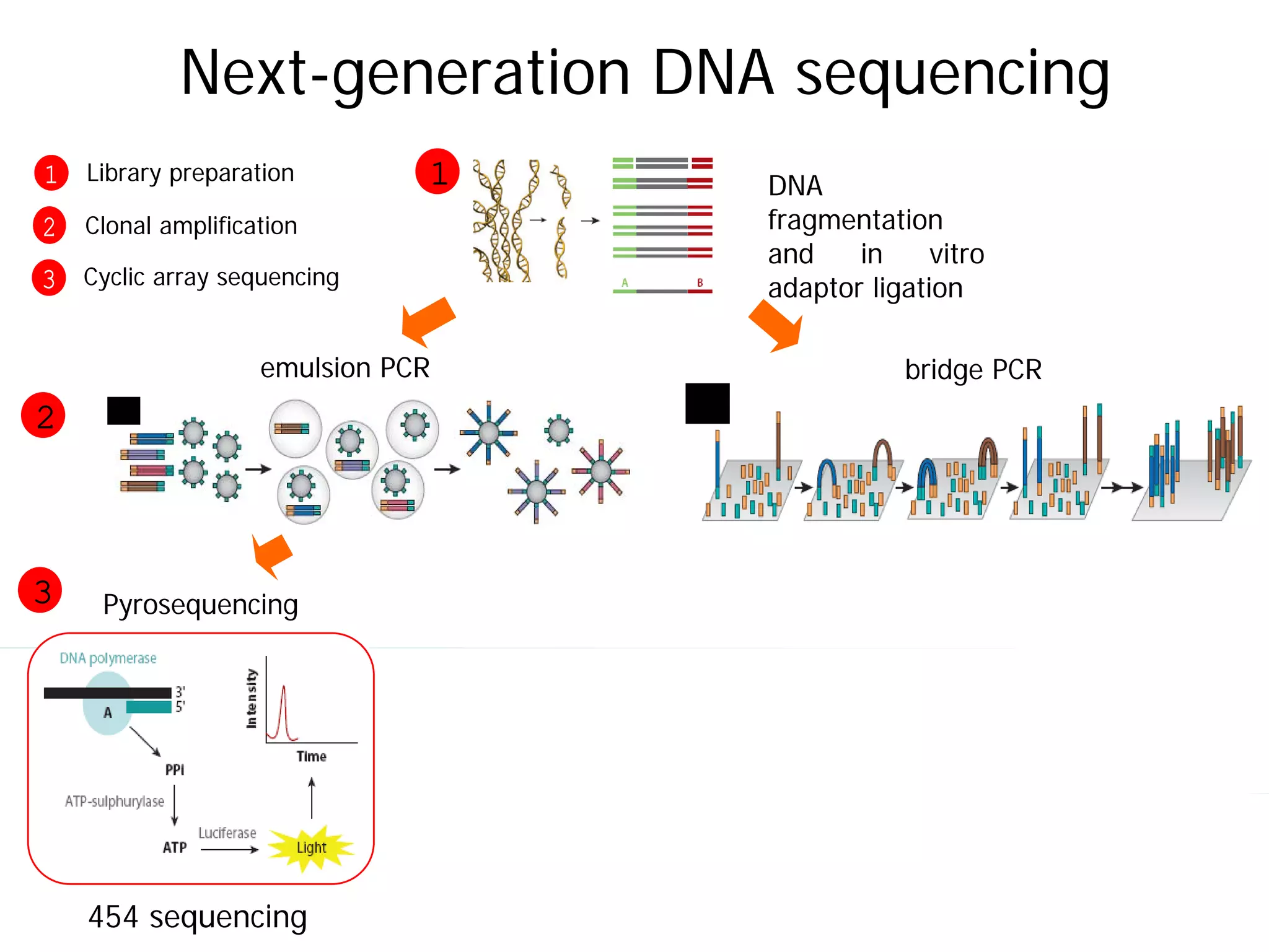
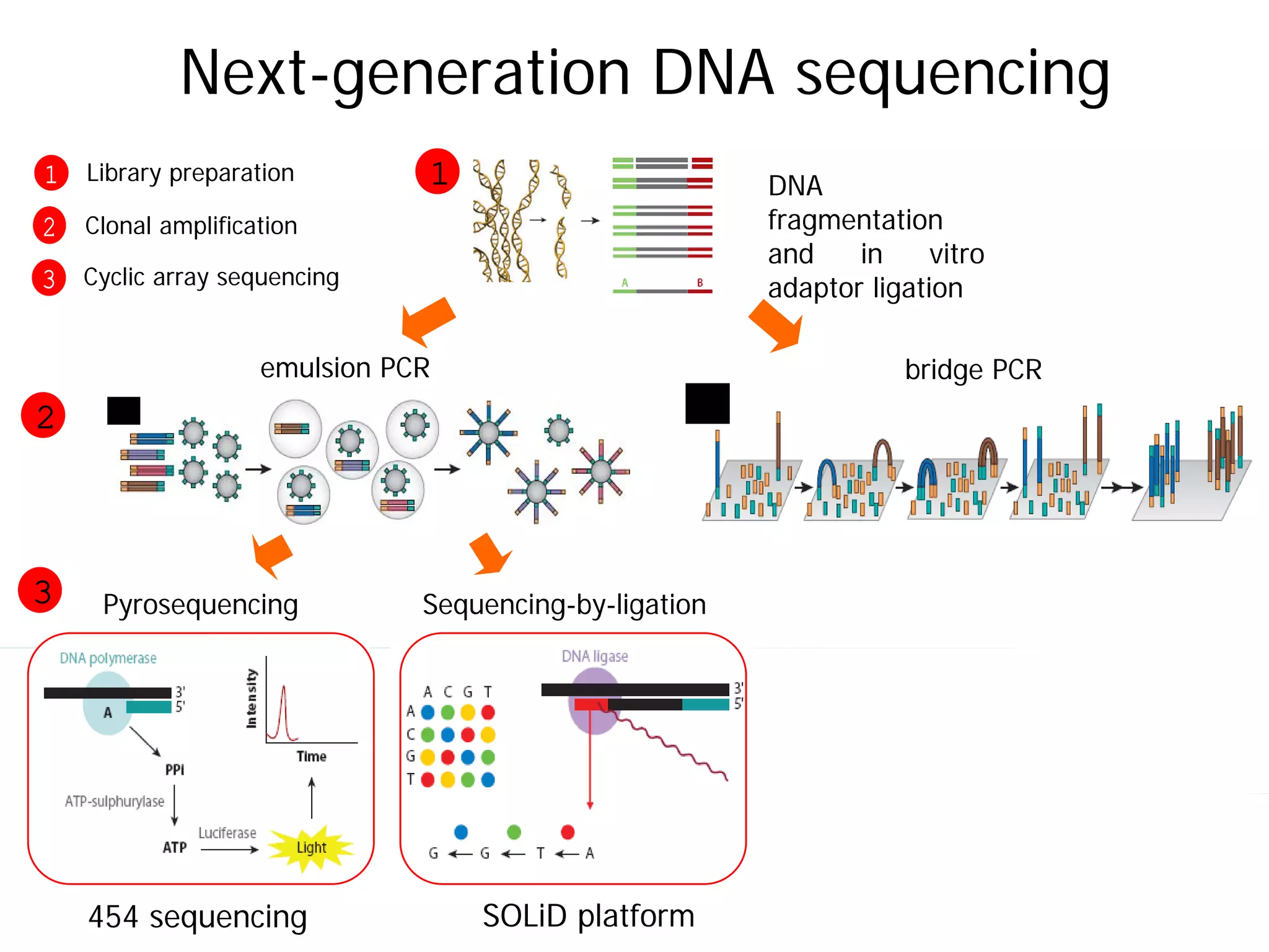
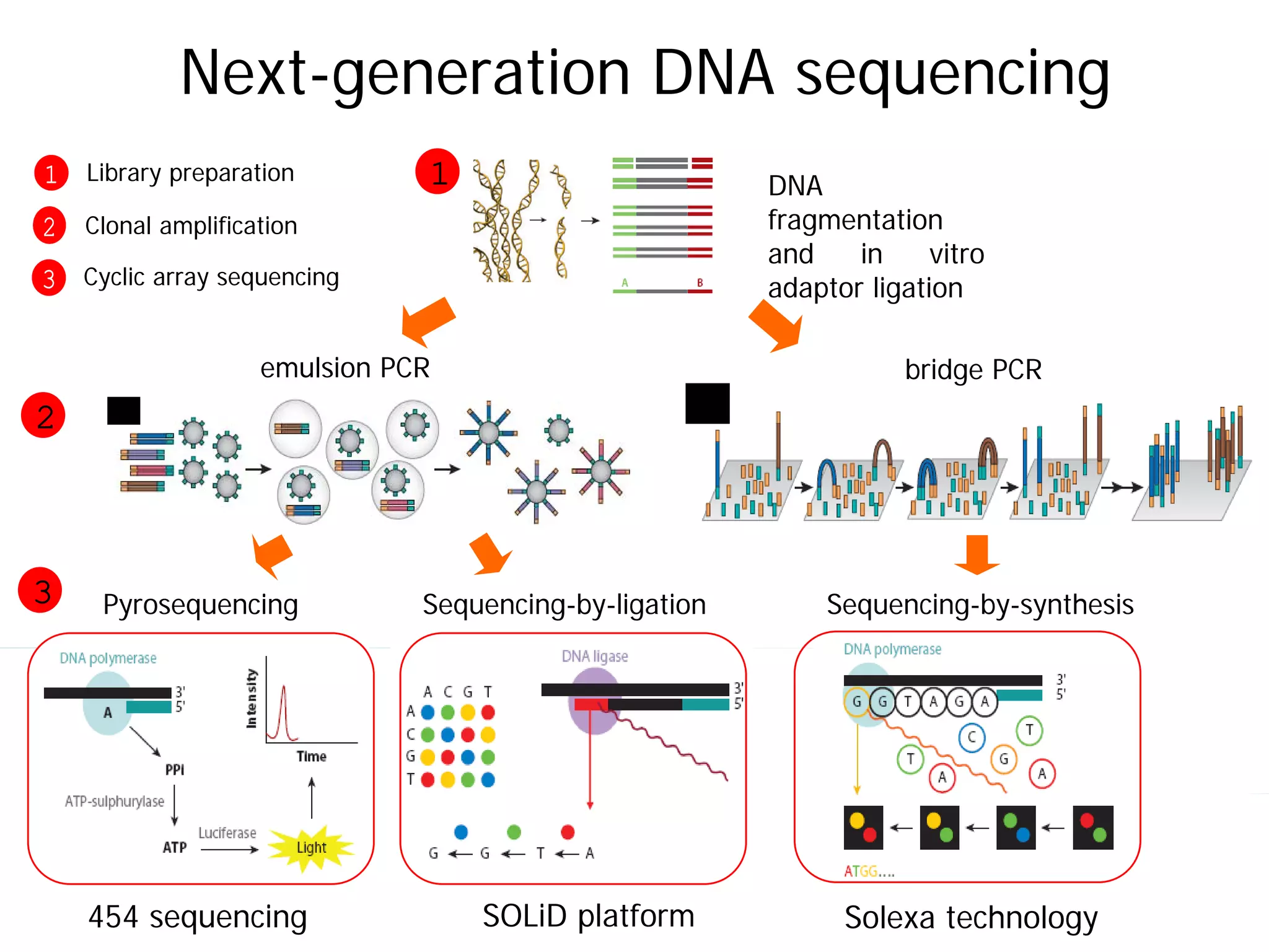
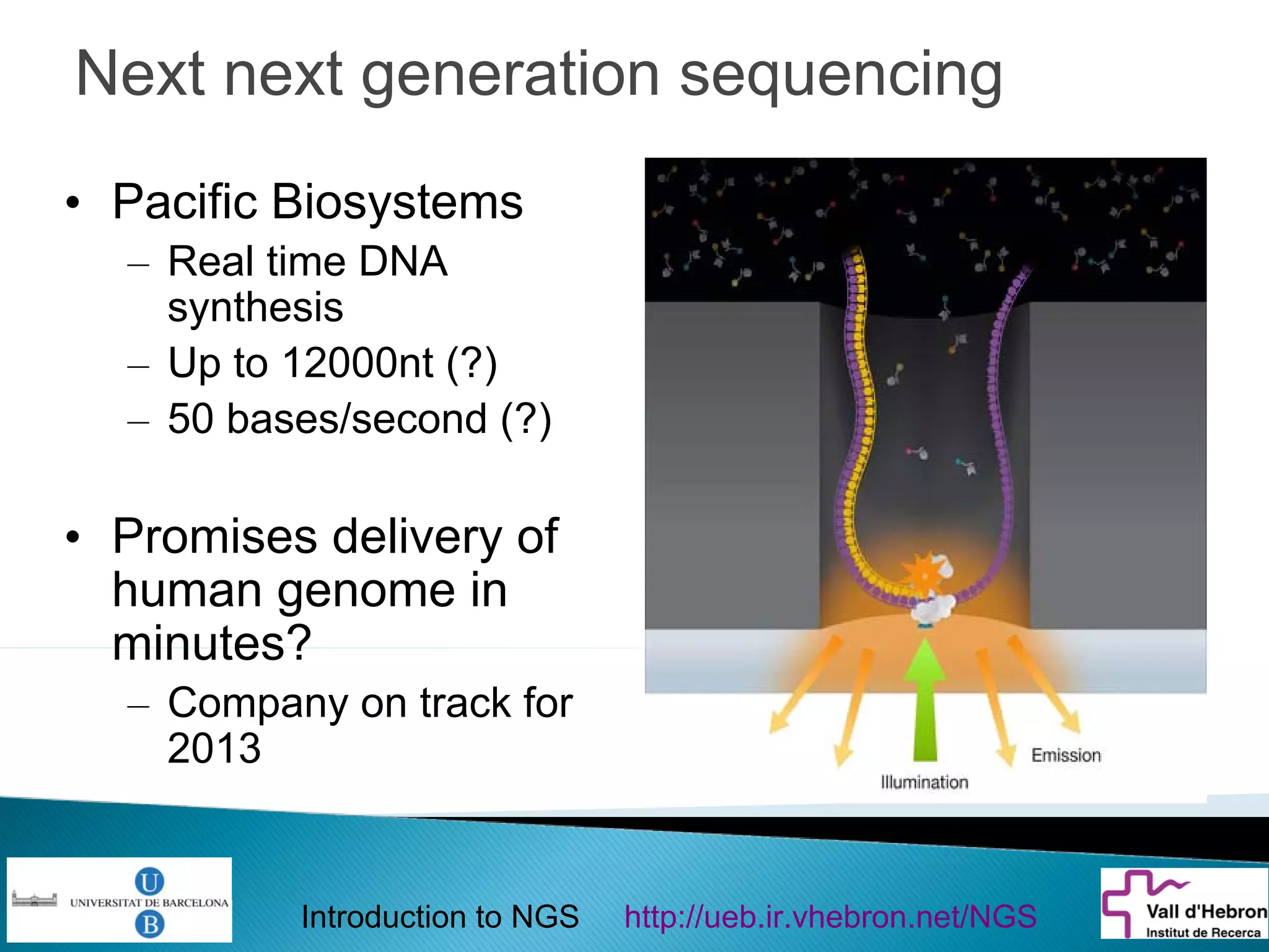
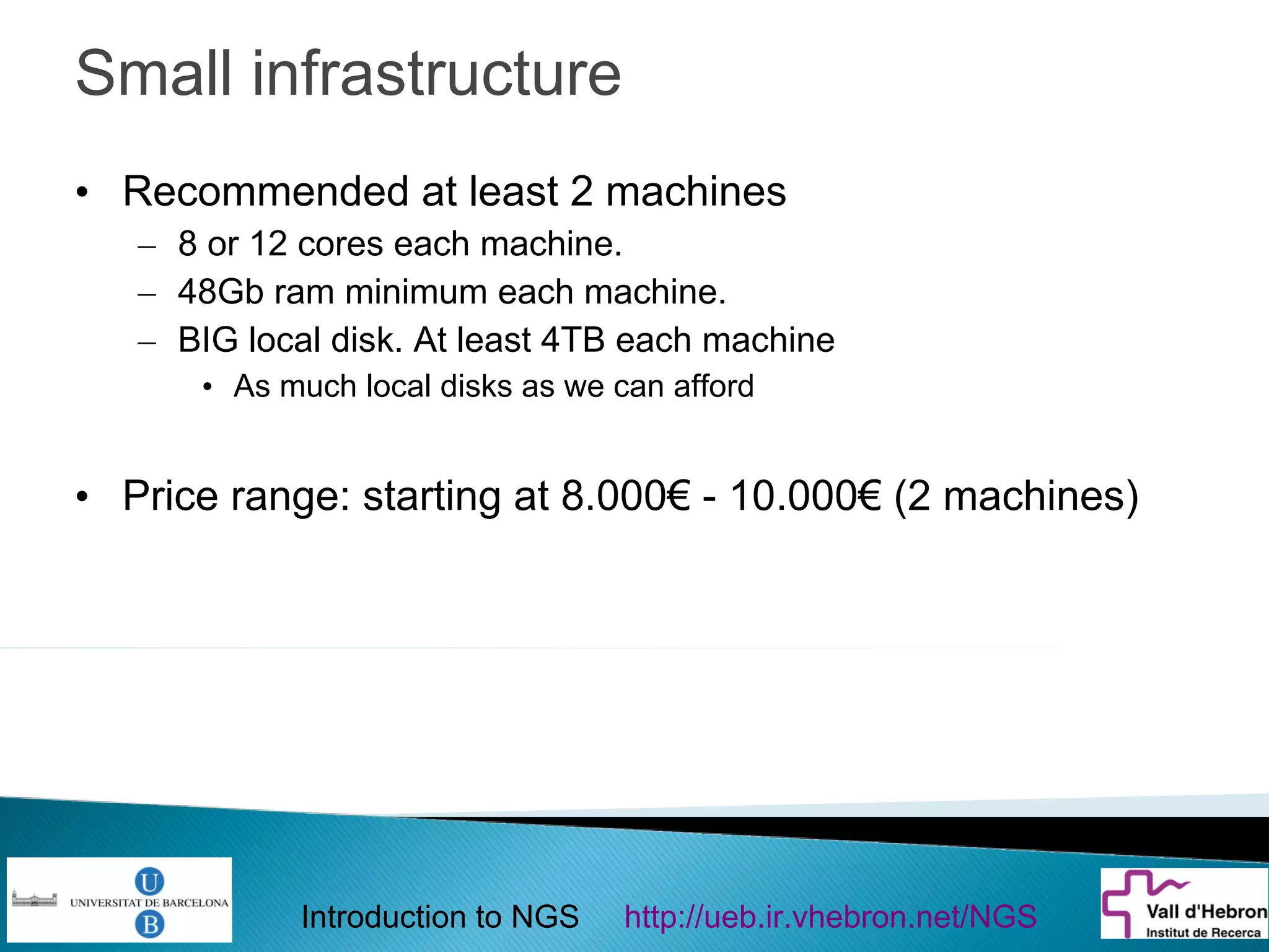
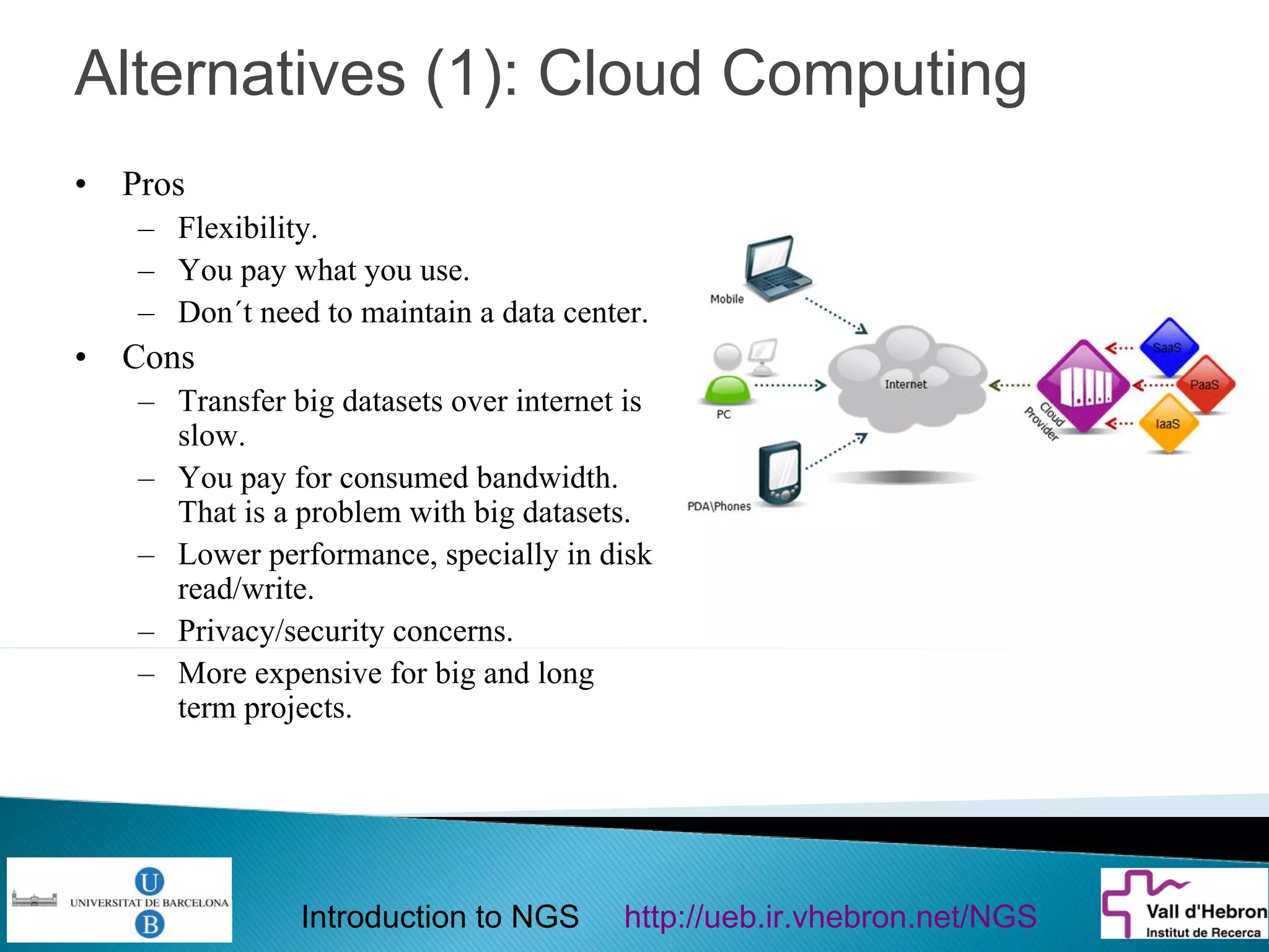
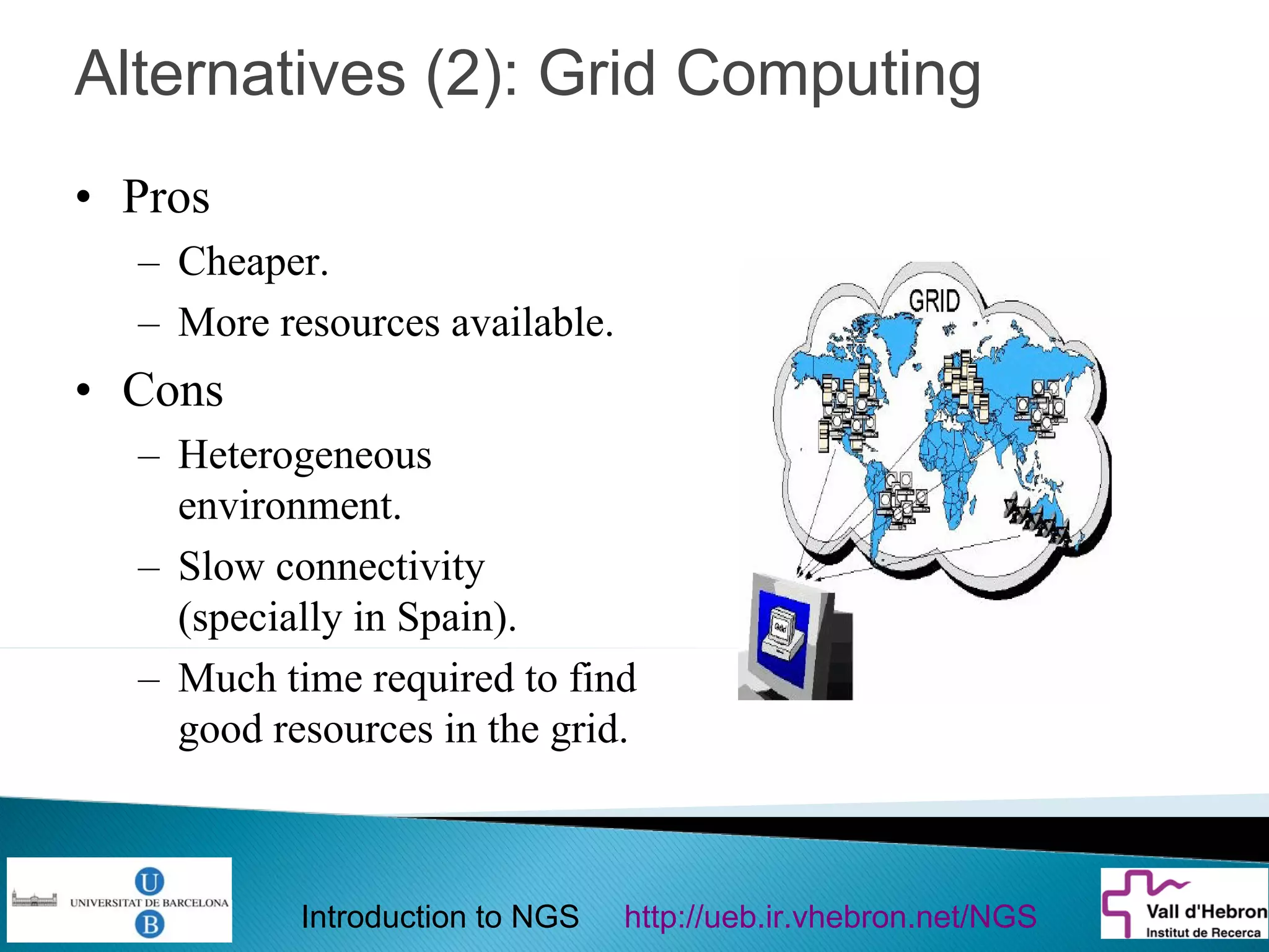
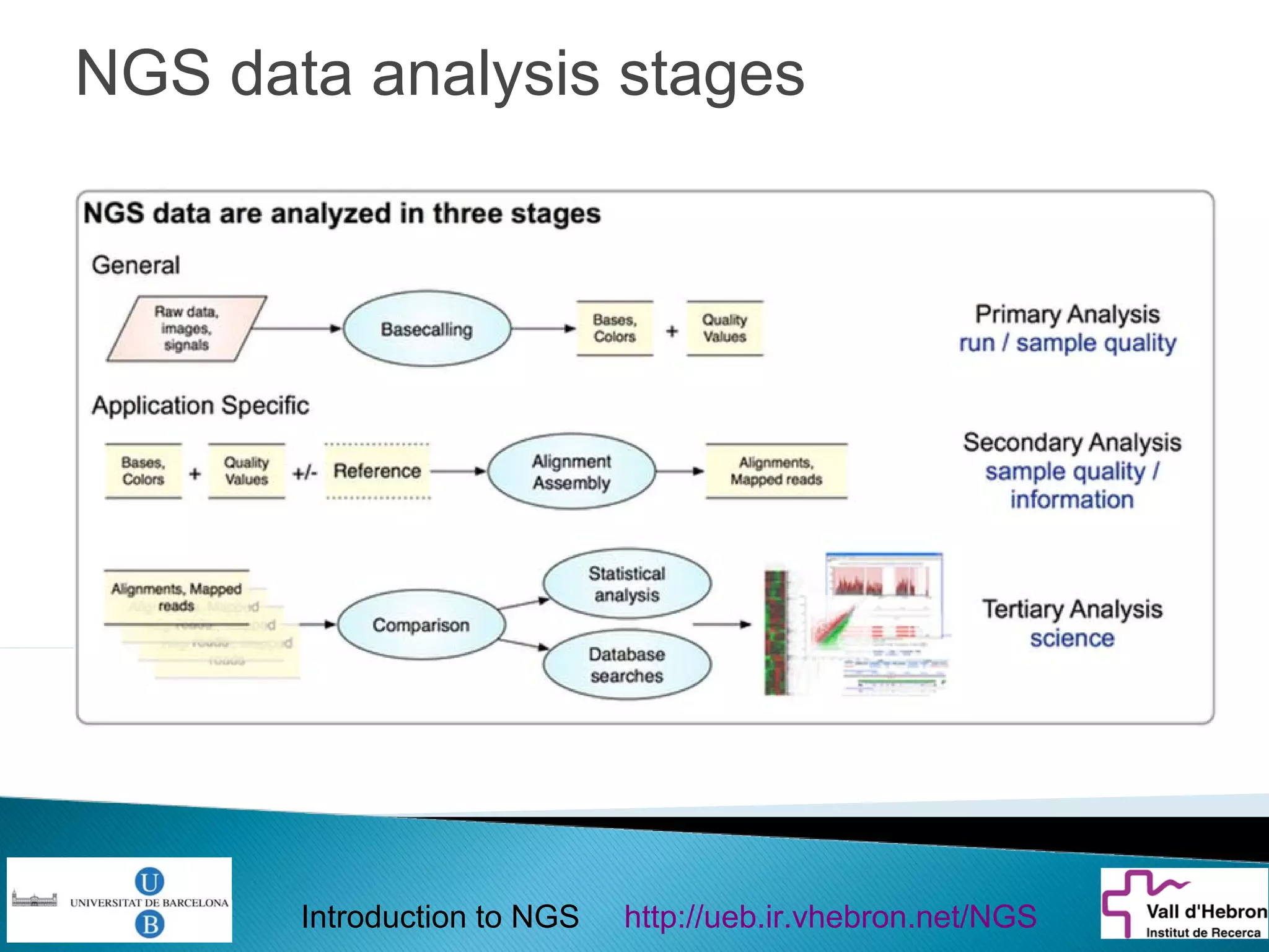
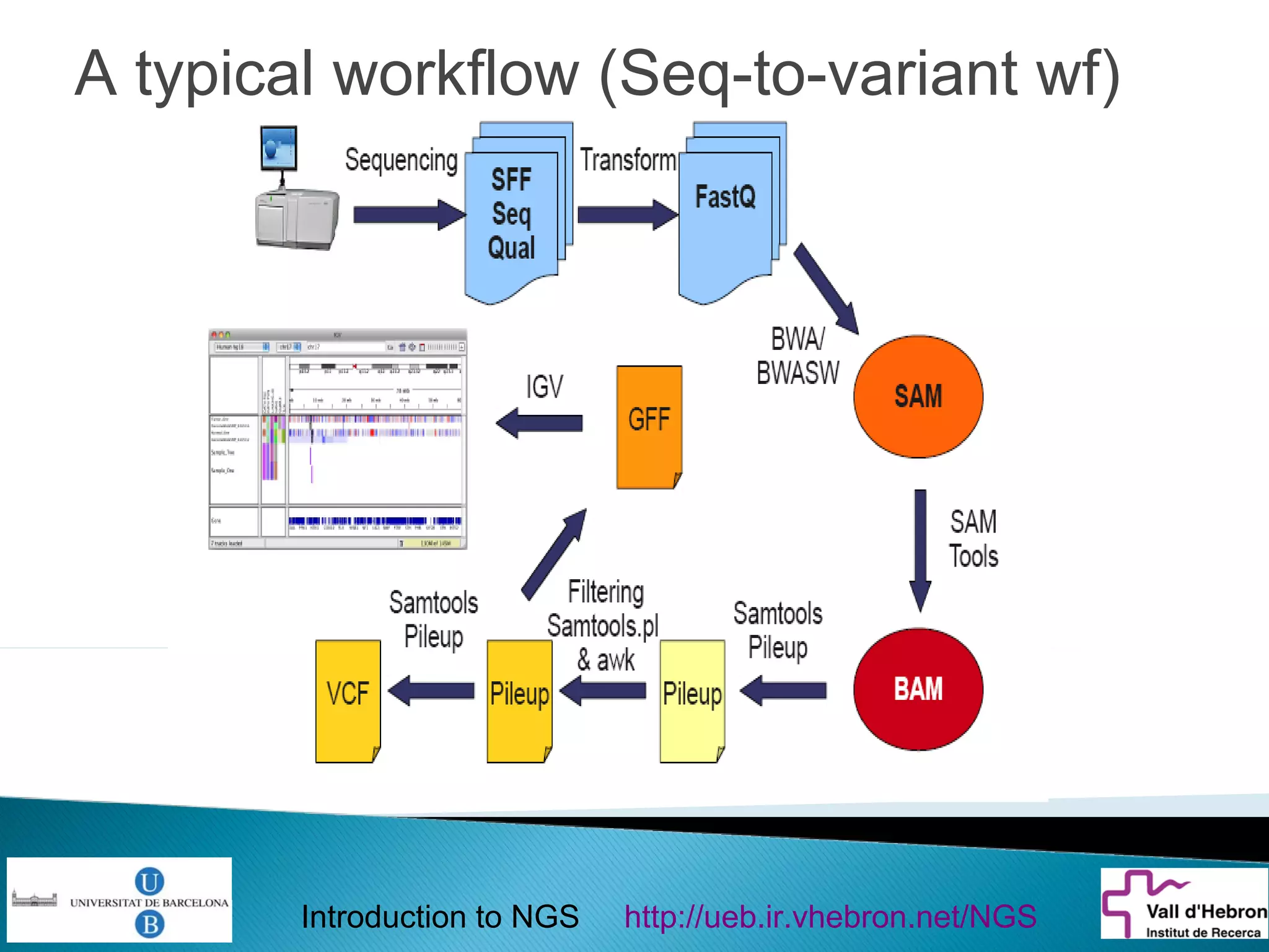
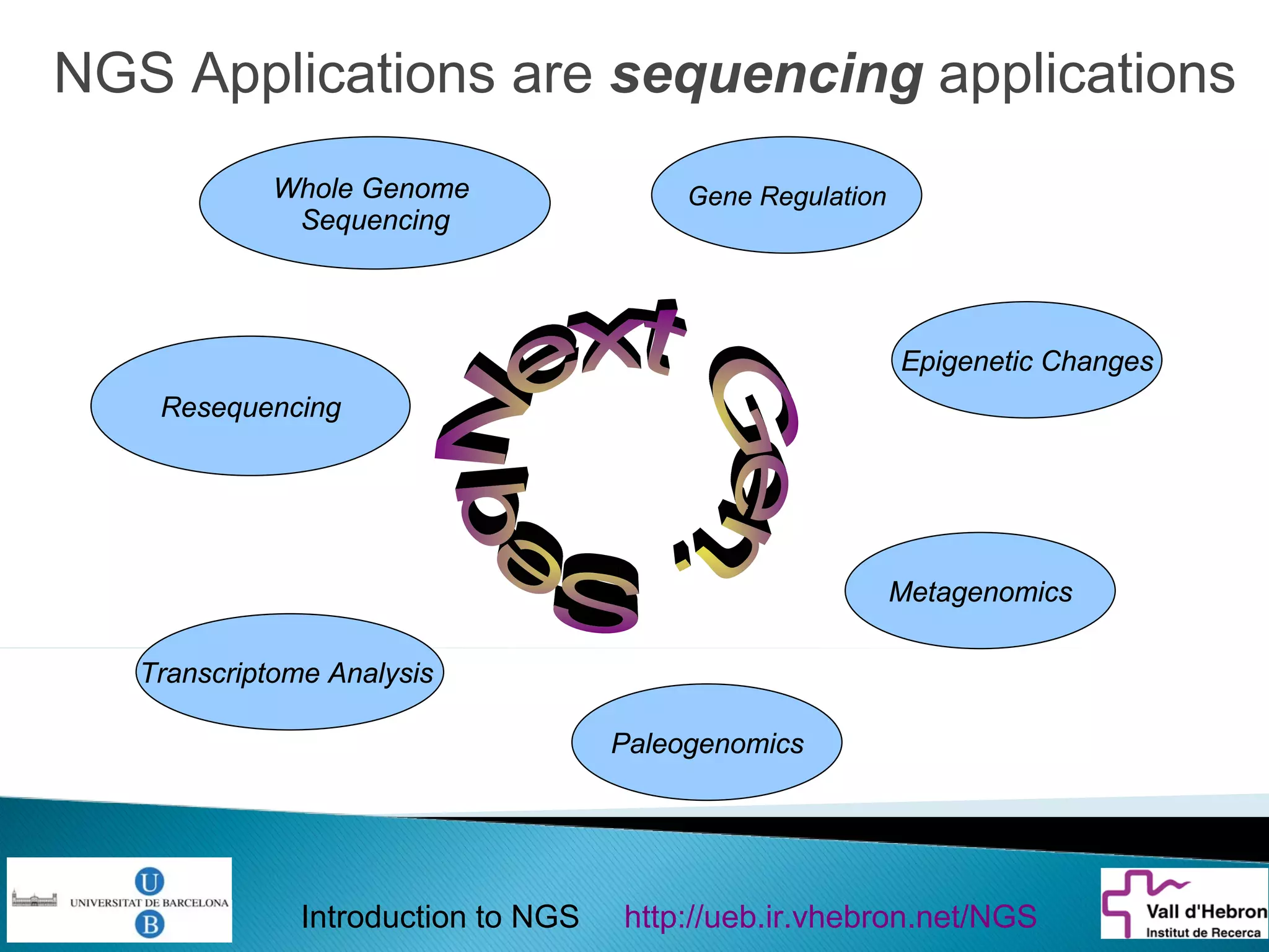
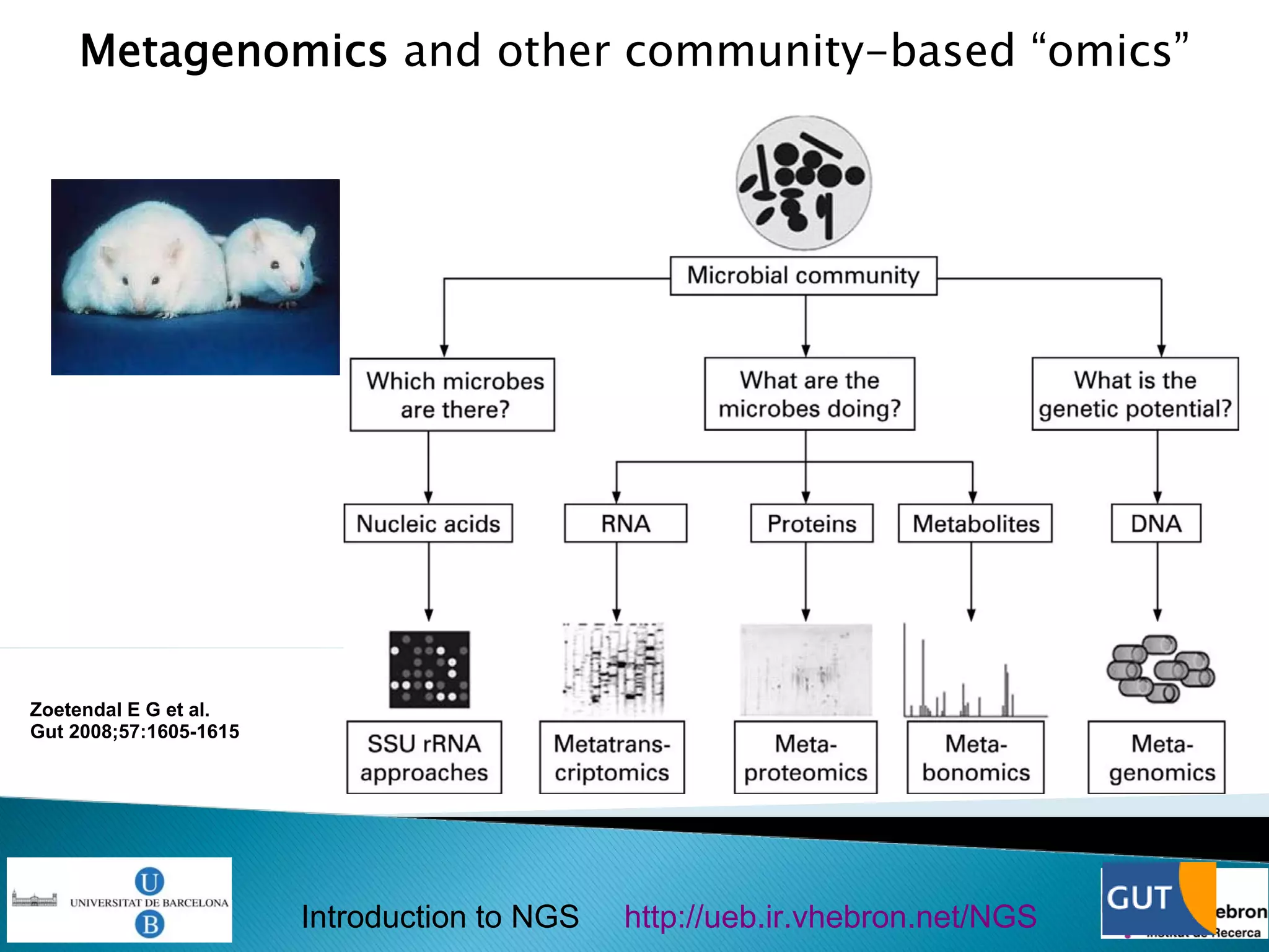
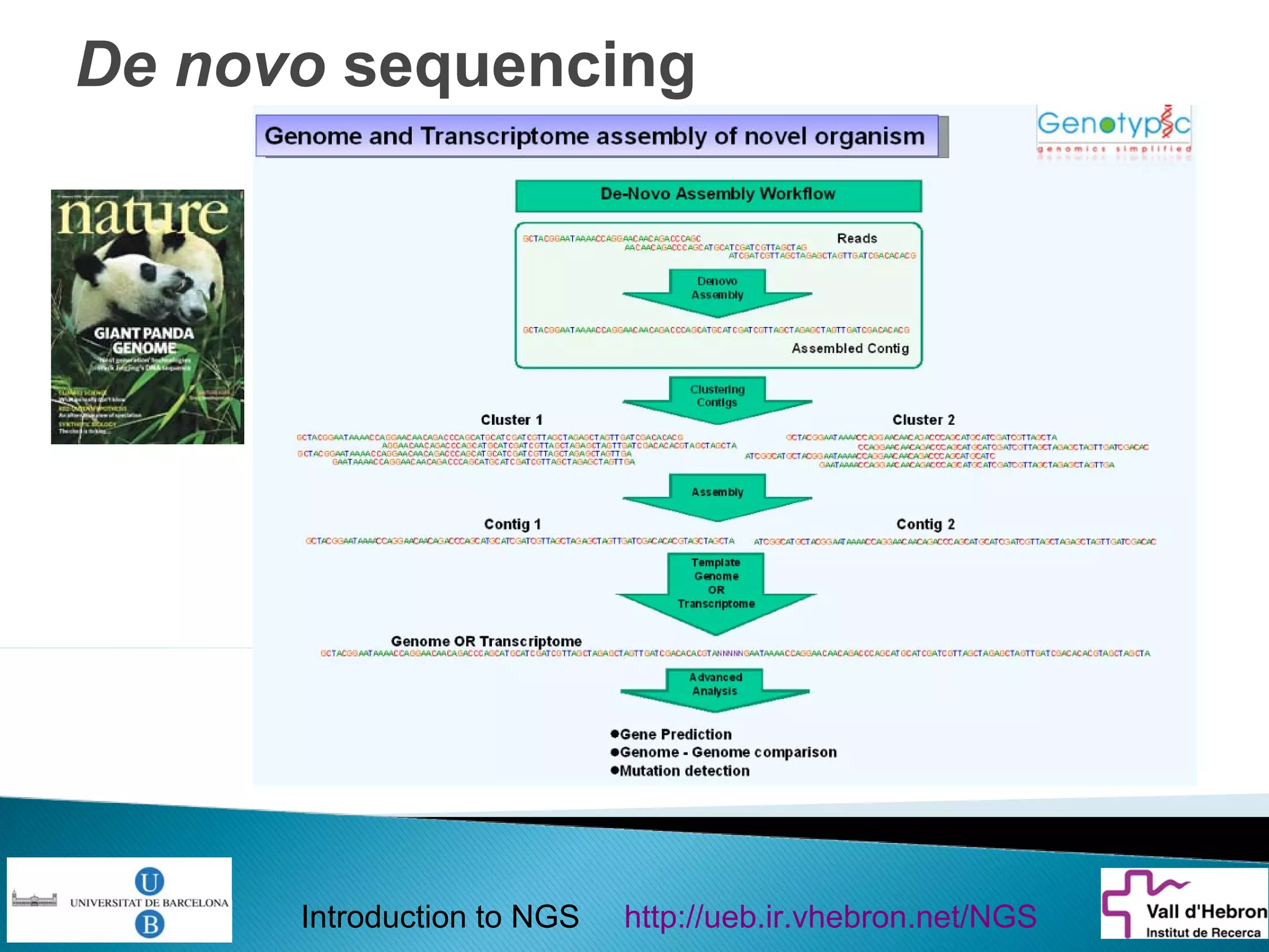
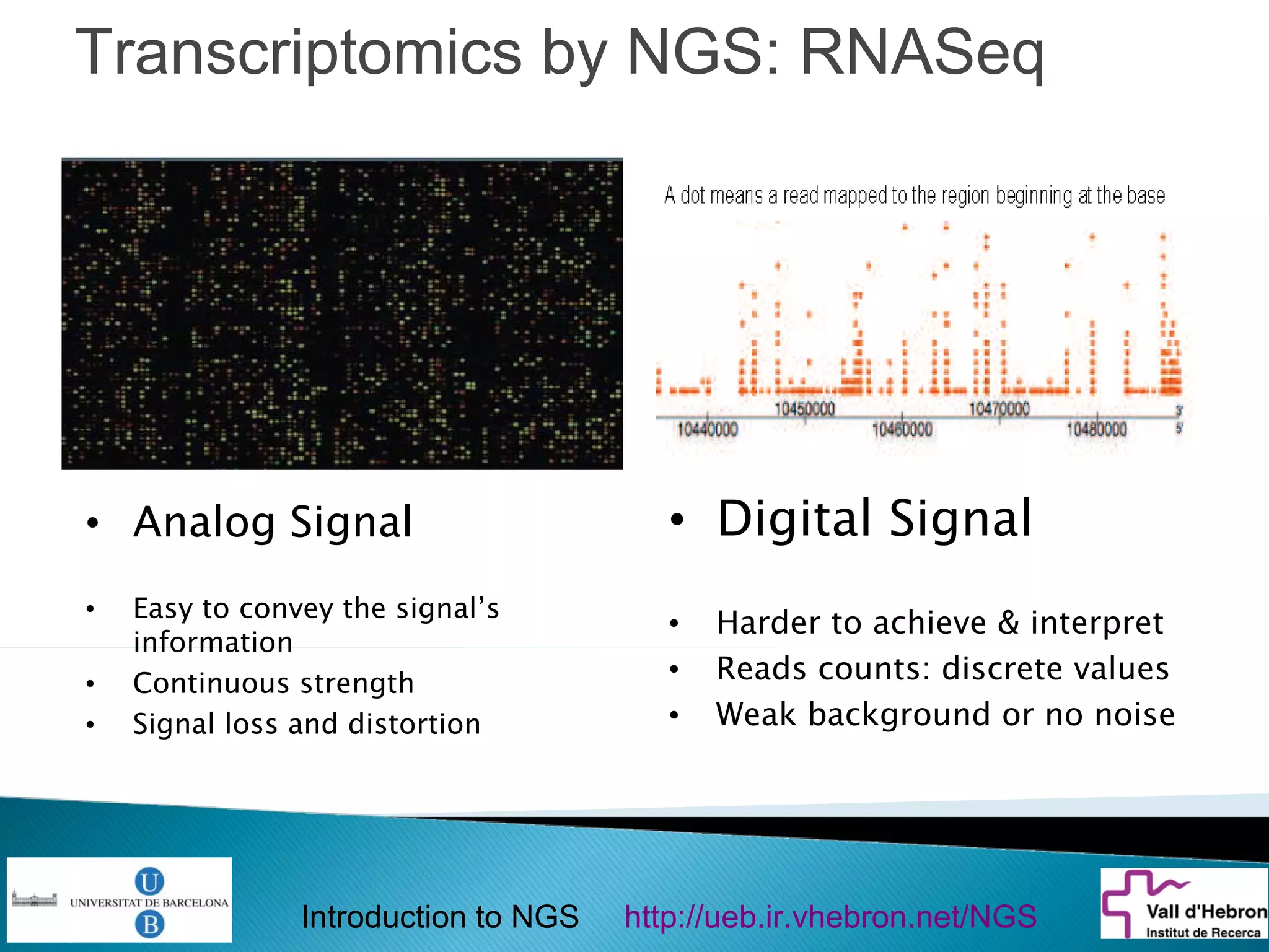
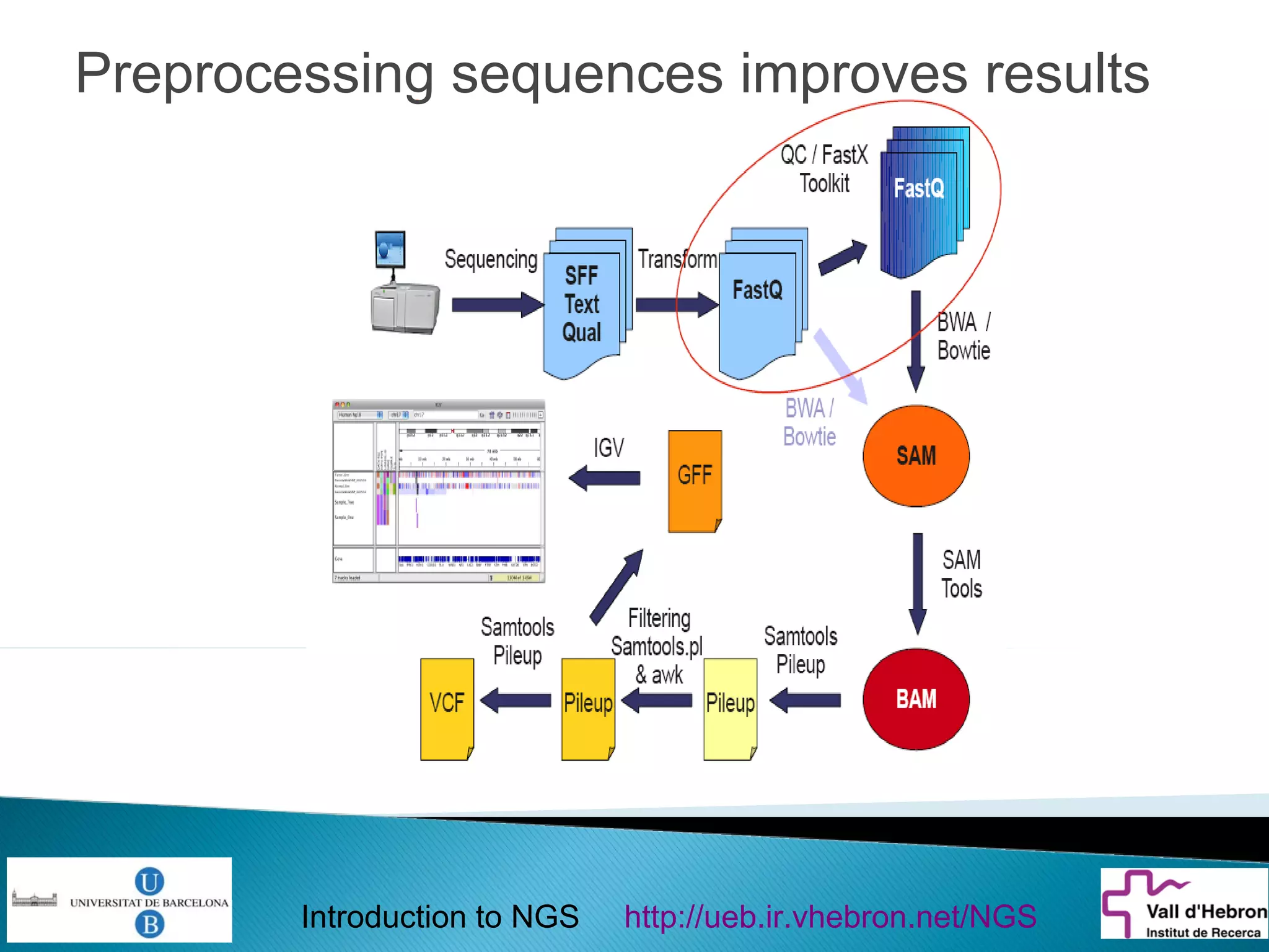
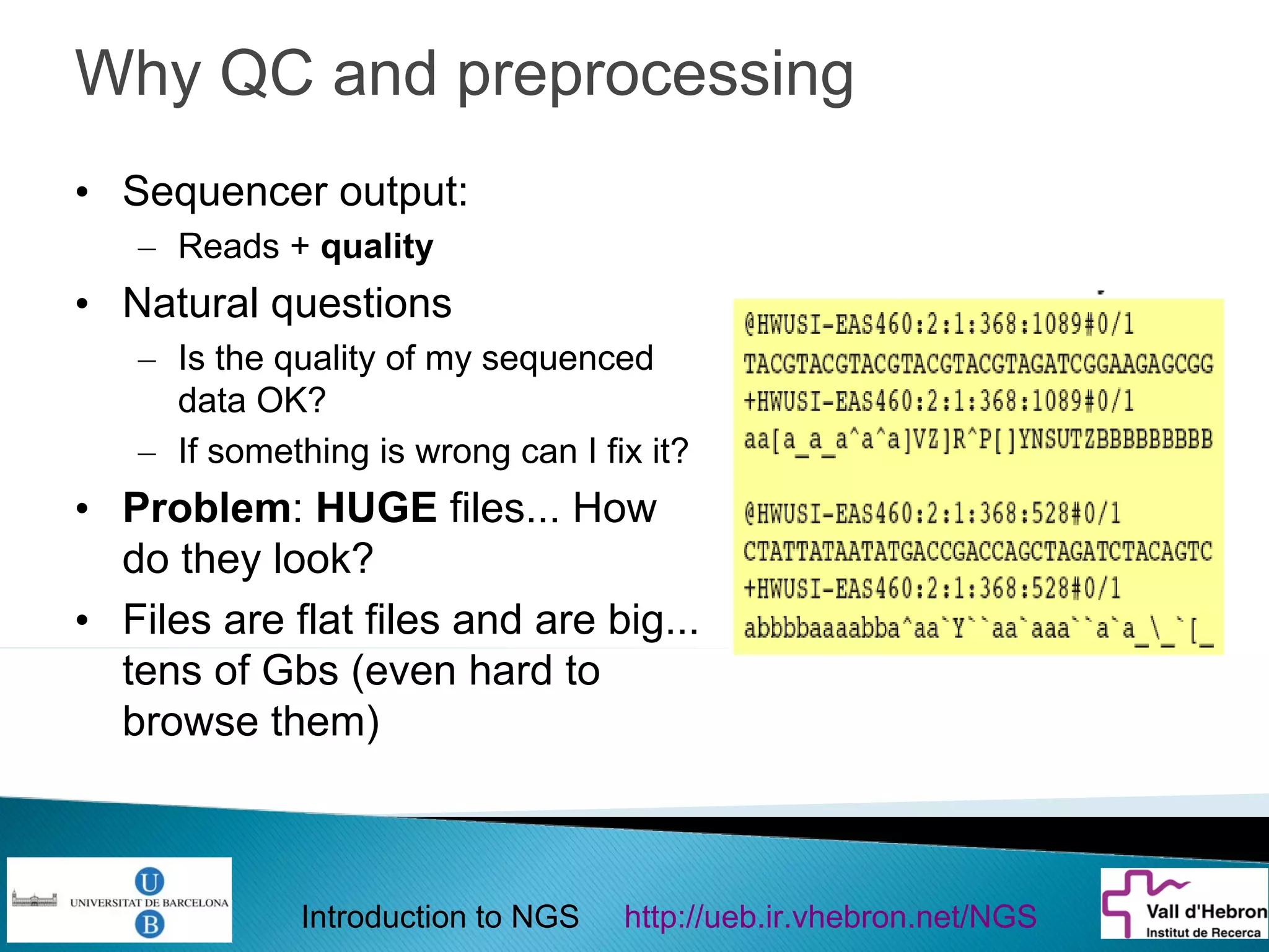
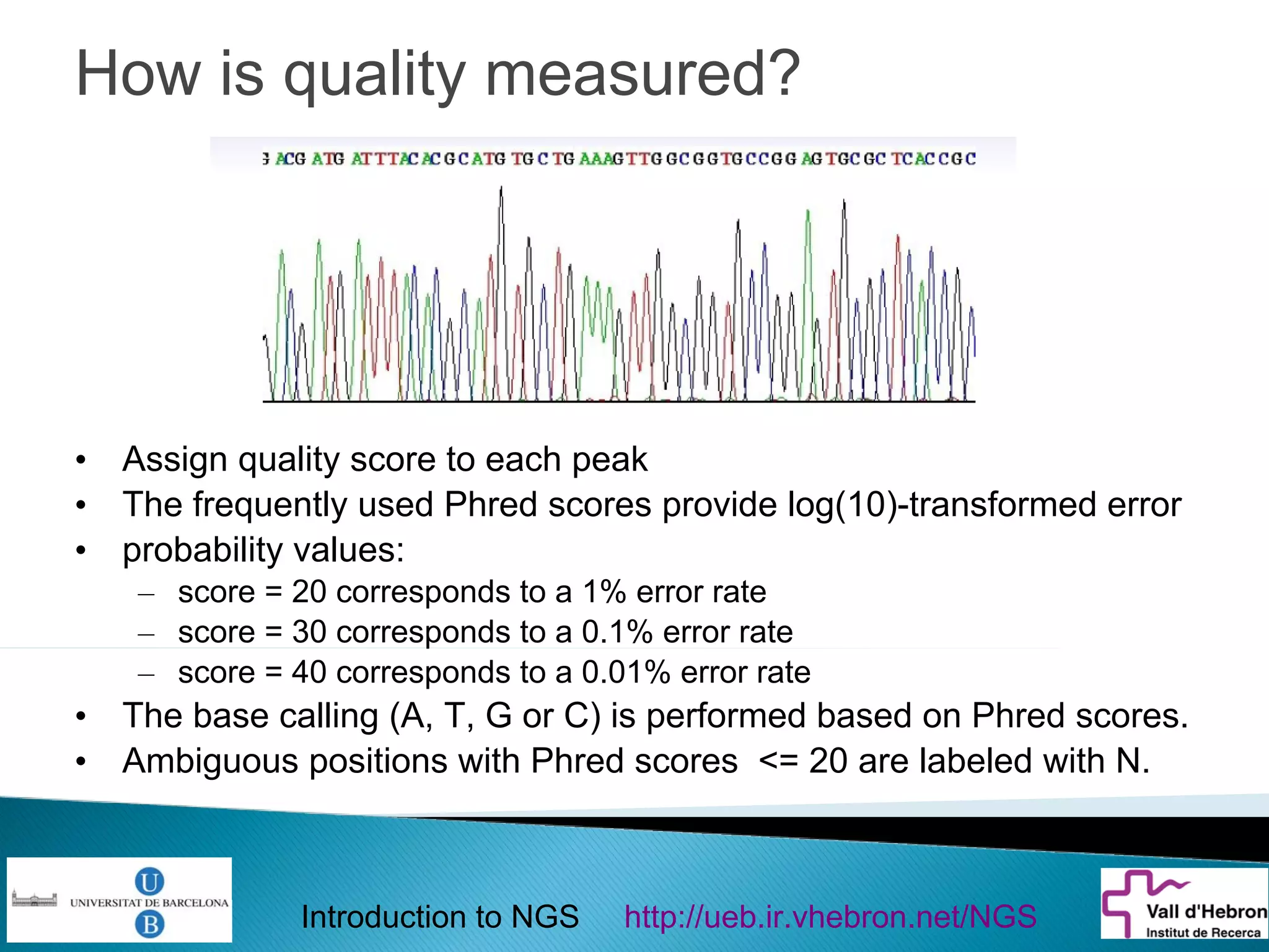
This document provides an introduction to next generation sequencing (NGS) technologies. It begins with an outline of topics to be covered, including the evolution of NGS technologies, their descriptions and comparisons, bioinformatics challenges of NGS data analysis, and some aspects of NGS data analysis workflows and tools. The document then delves into explanations of specific NGS platforms, their performance characteristics, and the sequencing processes. It discusses the large computational infrastructure and data management needs of NGS, as well as quality control, preprocessing of NGS data, and popular analysis tools and workflows.