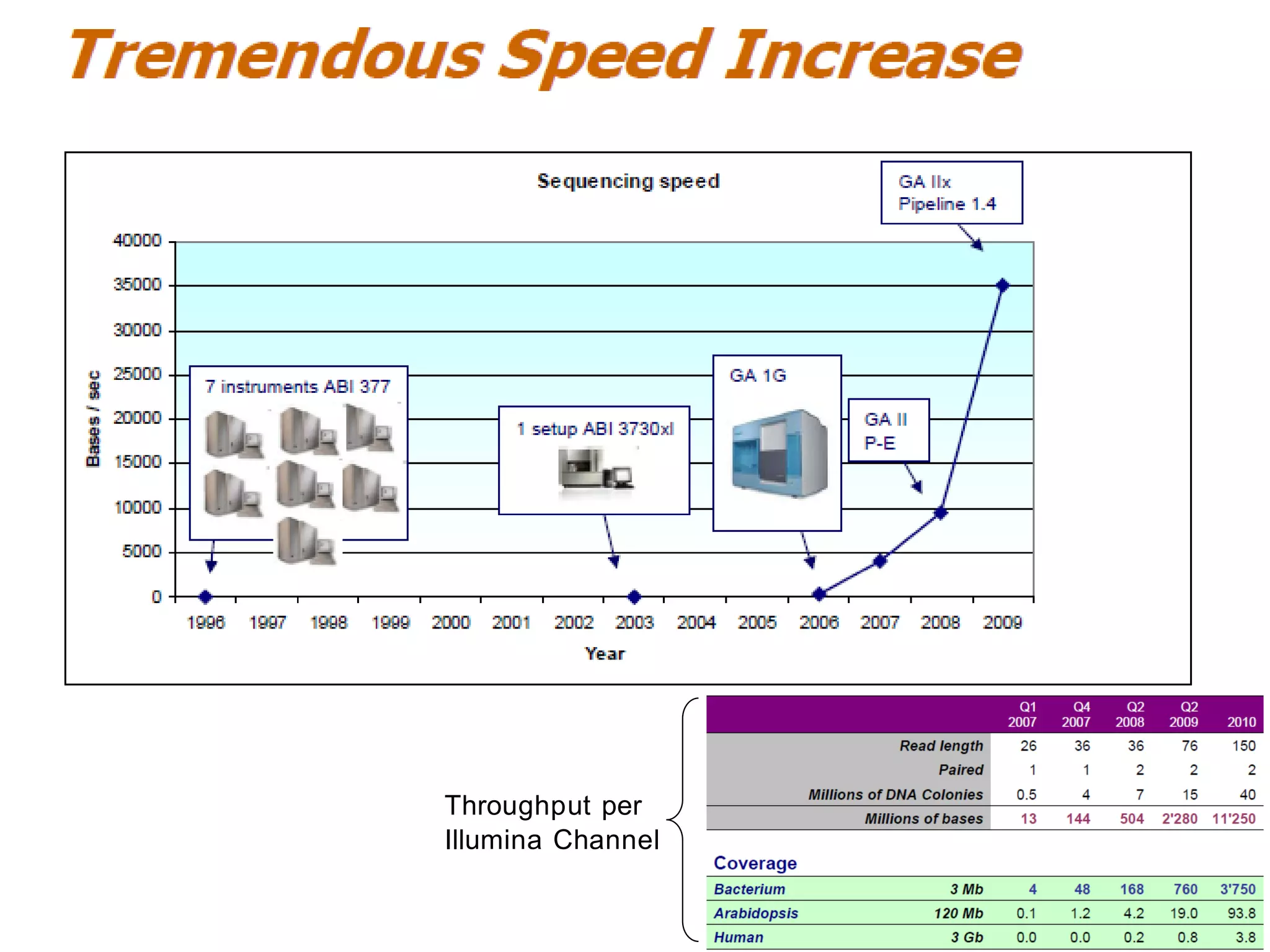
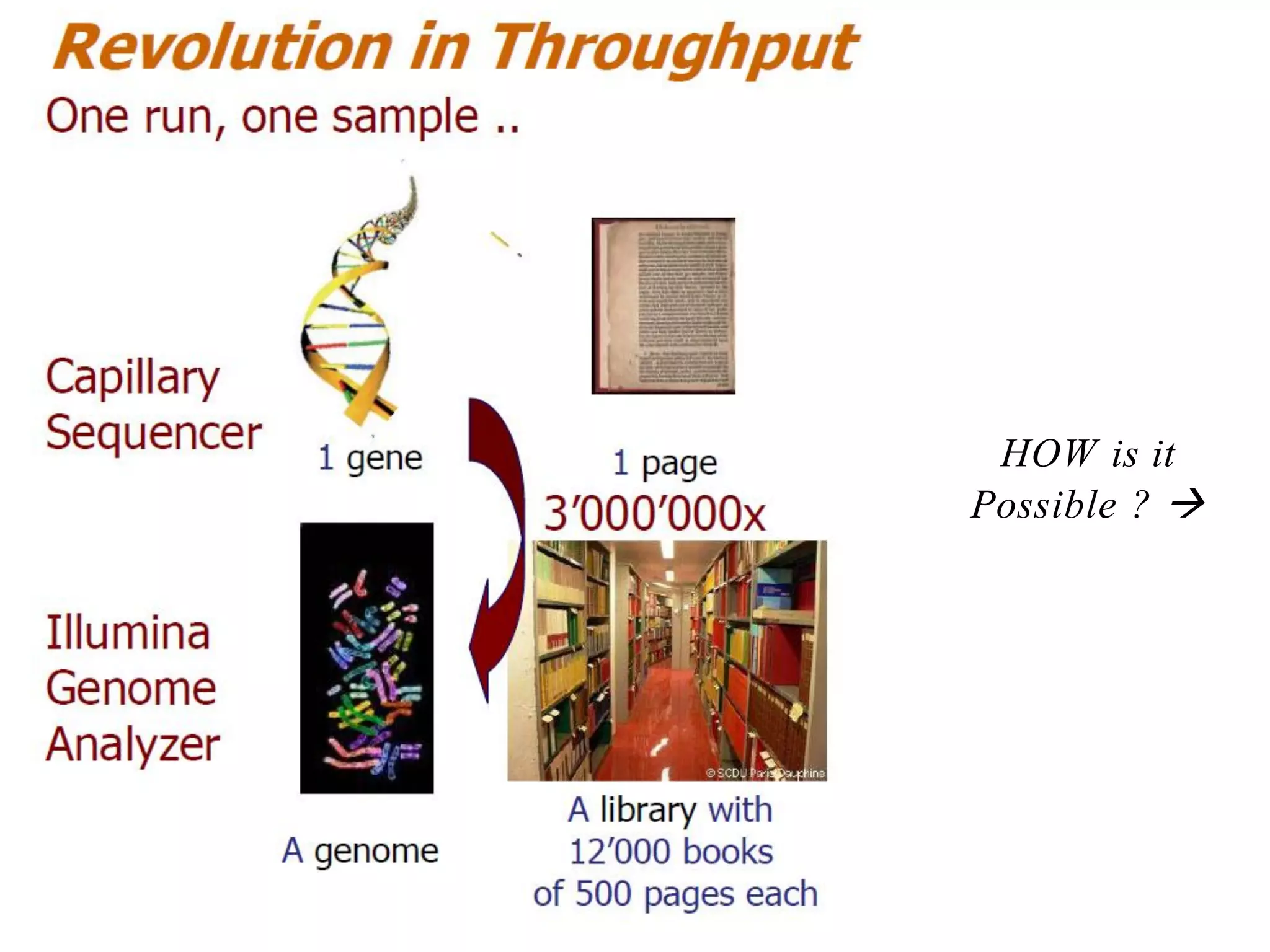
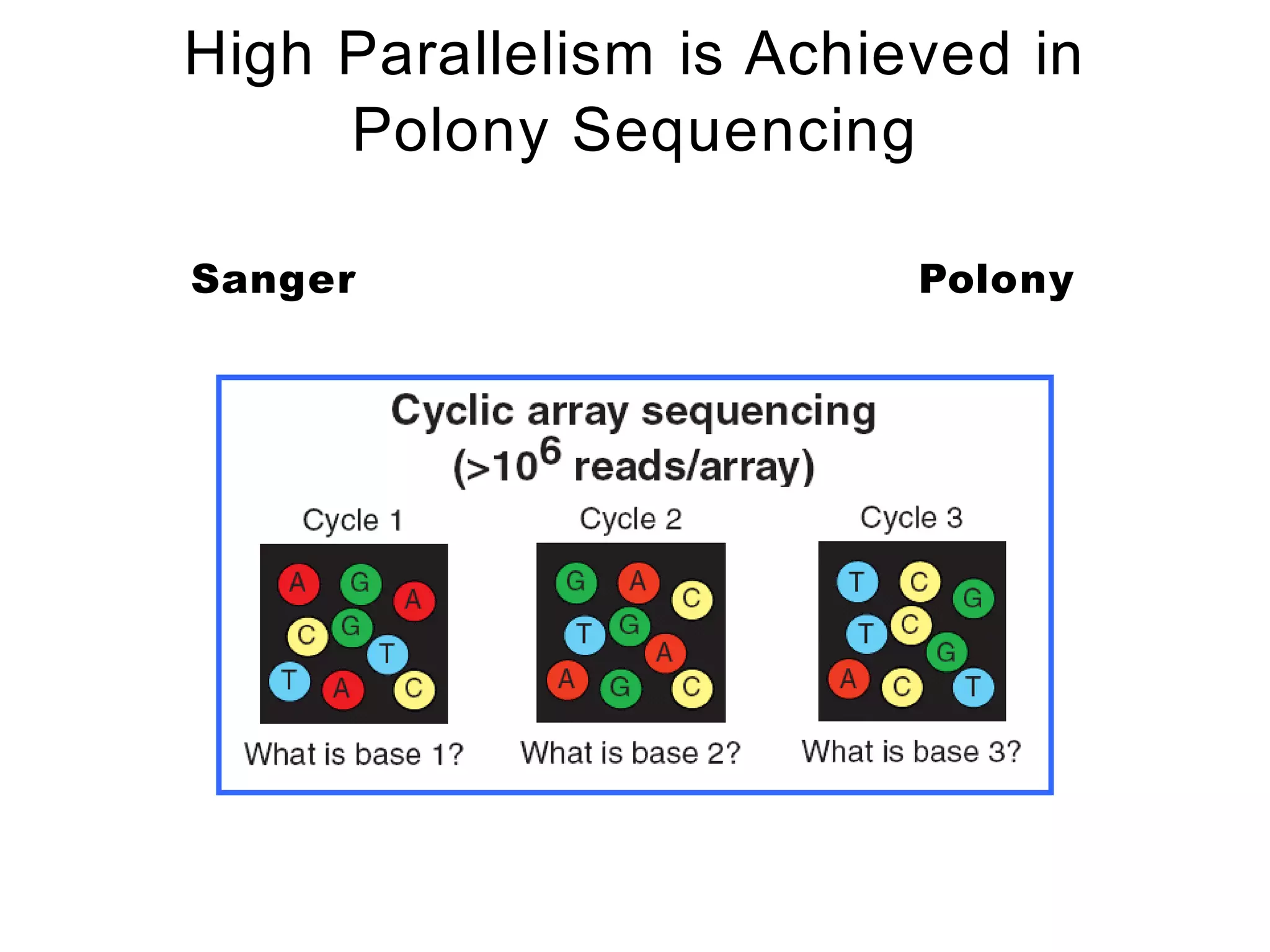
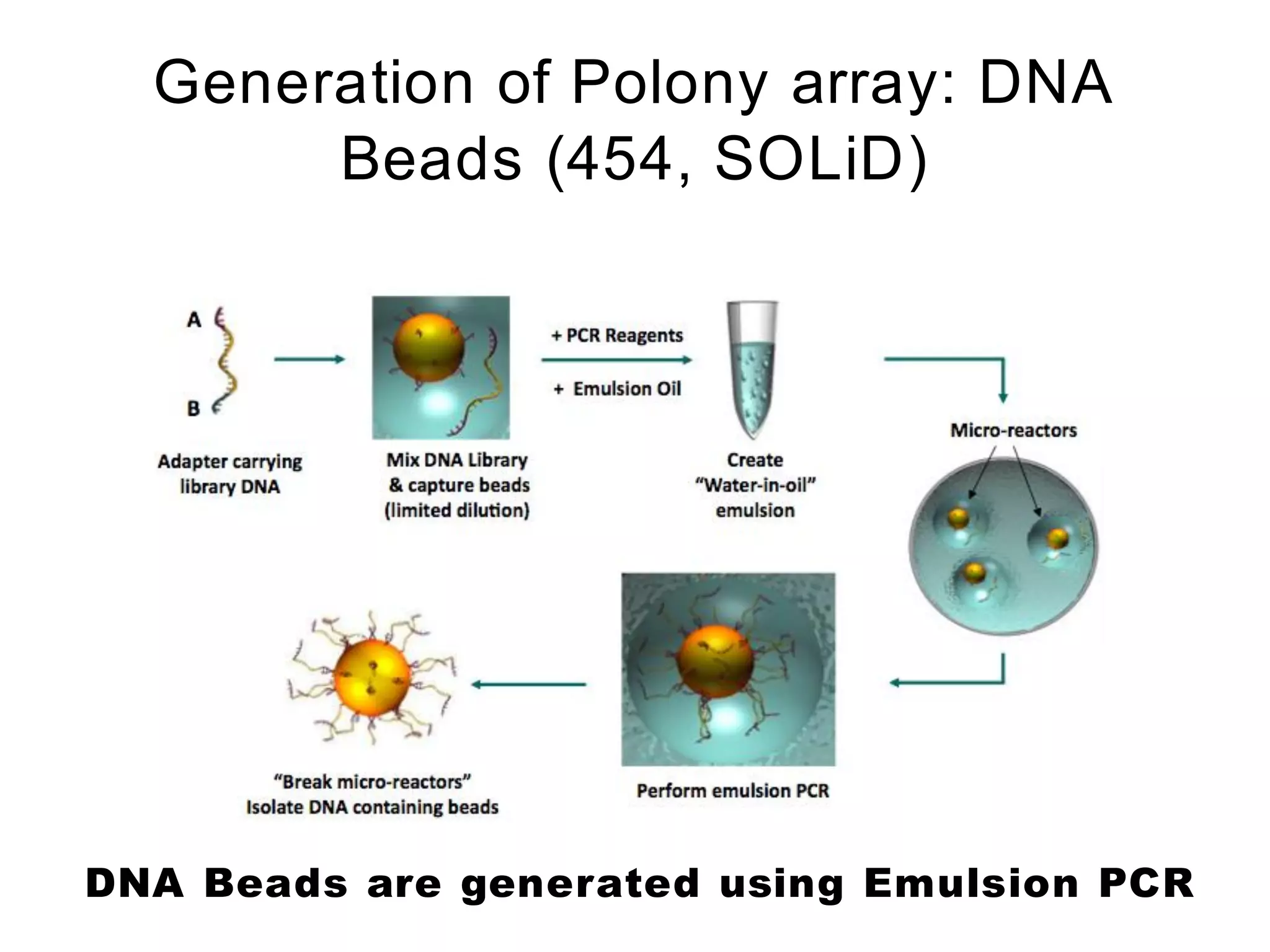
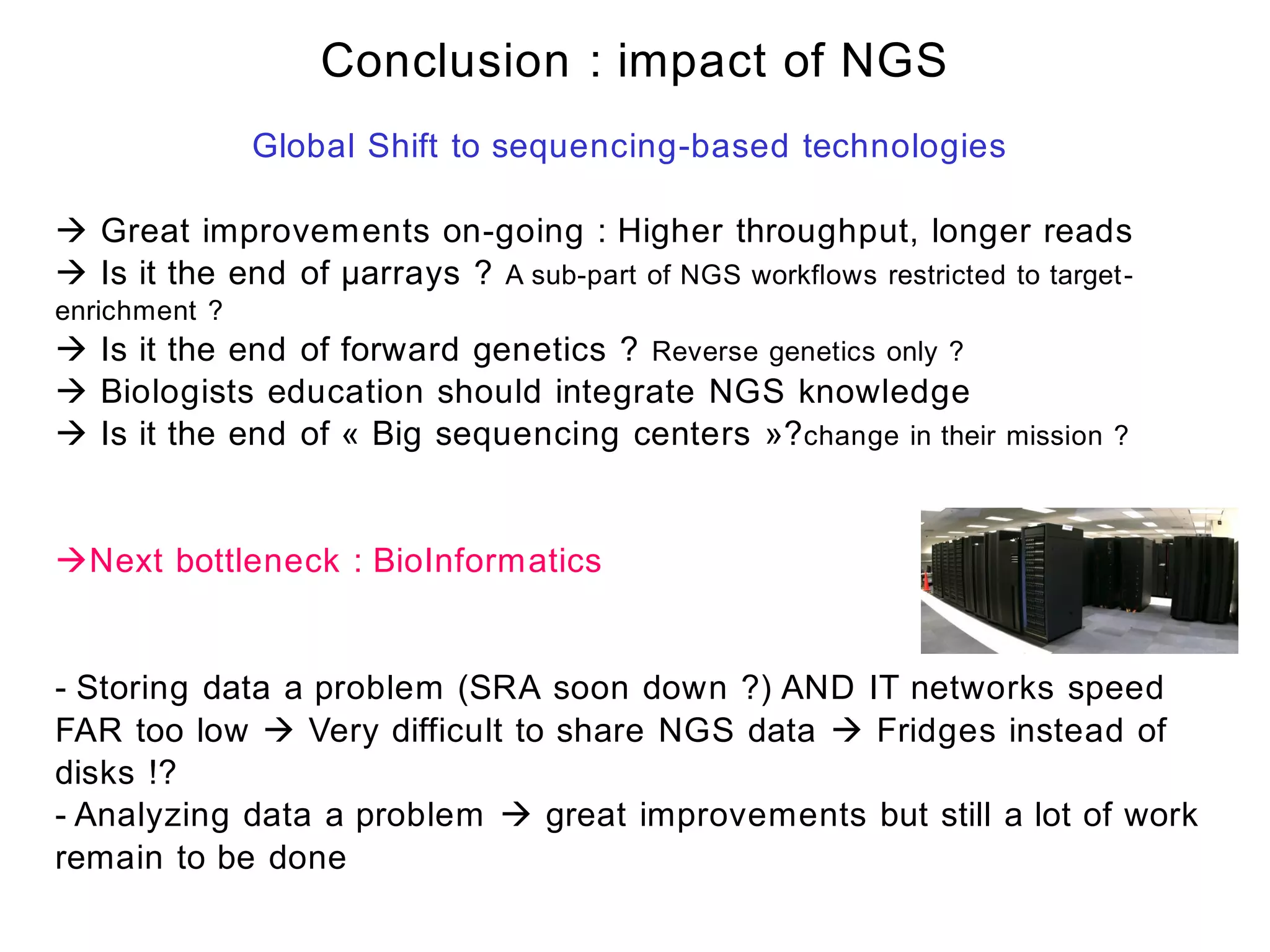
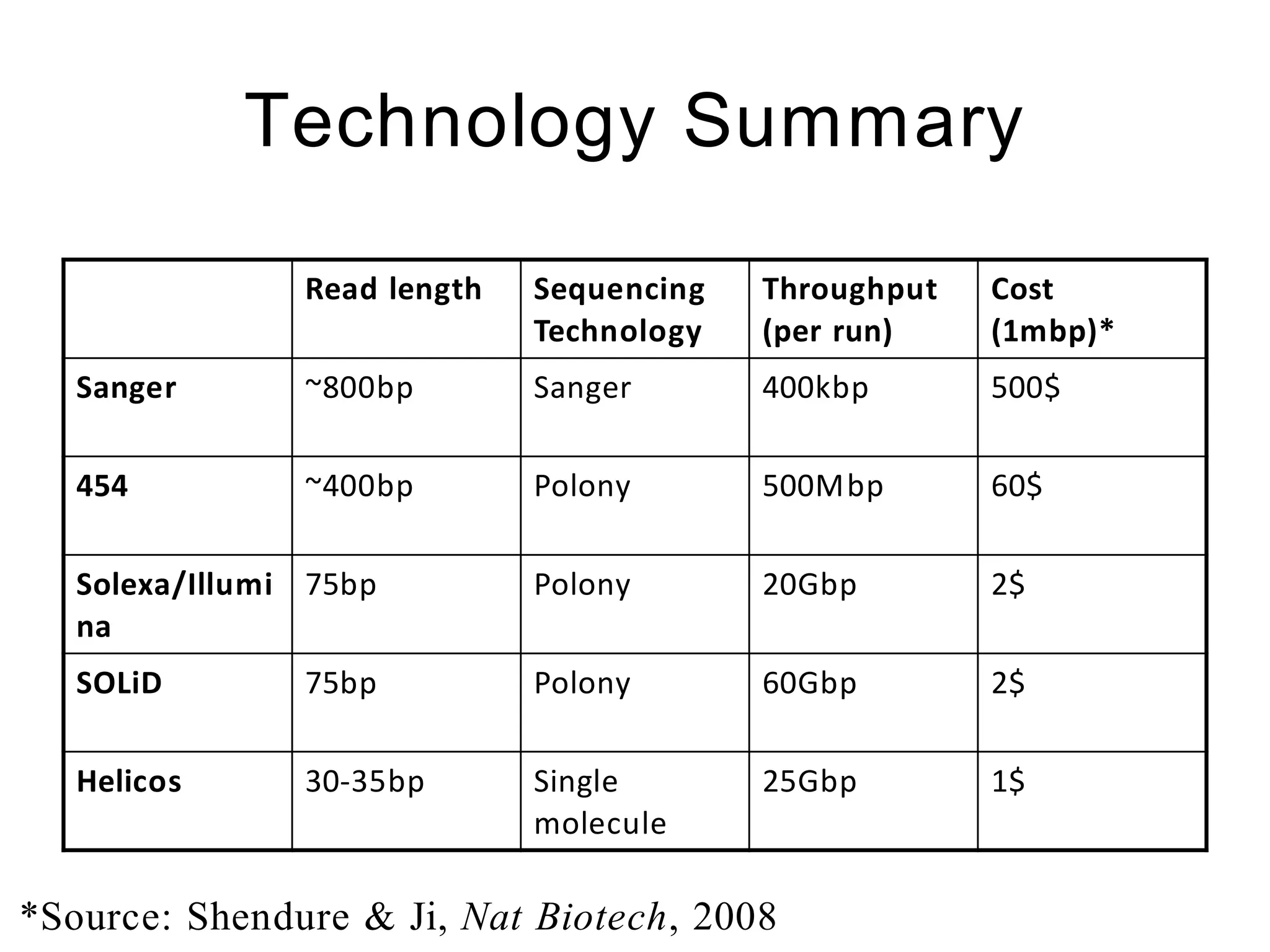
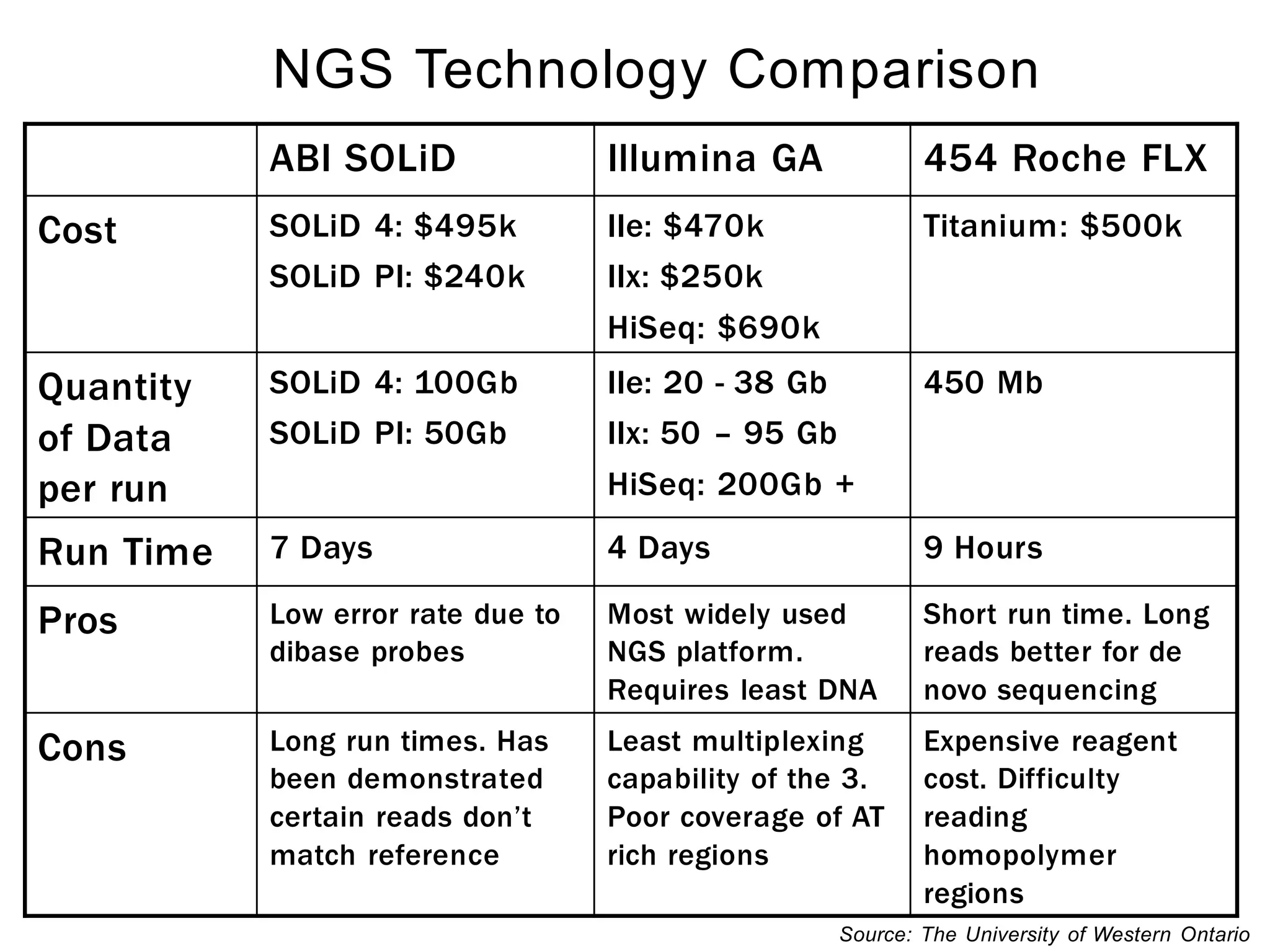
The document provides a comprehensive introduction to next-generation sequencing (NGS), discussing its evolution from Sanger sequencing and detailing various NGS technologies and their applications. Key topics include workflow processes, assembly strategies, sequencing techniques from multiple companies, and the impact of NGS on genomics and personalized medicine. It highlights the advantages and limitations of different NGS platforms while emphasizing the growing importance of bioinformatics in managing and analyzing sequencing data.
![Plan
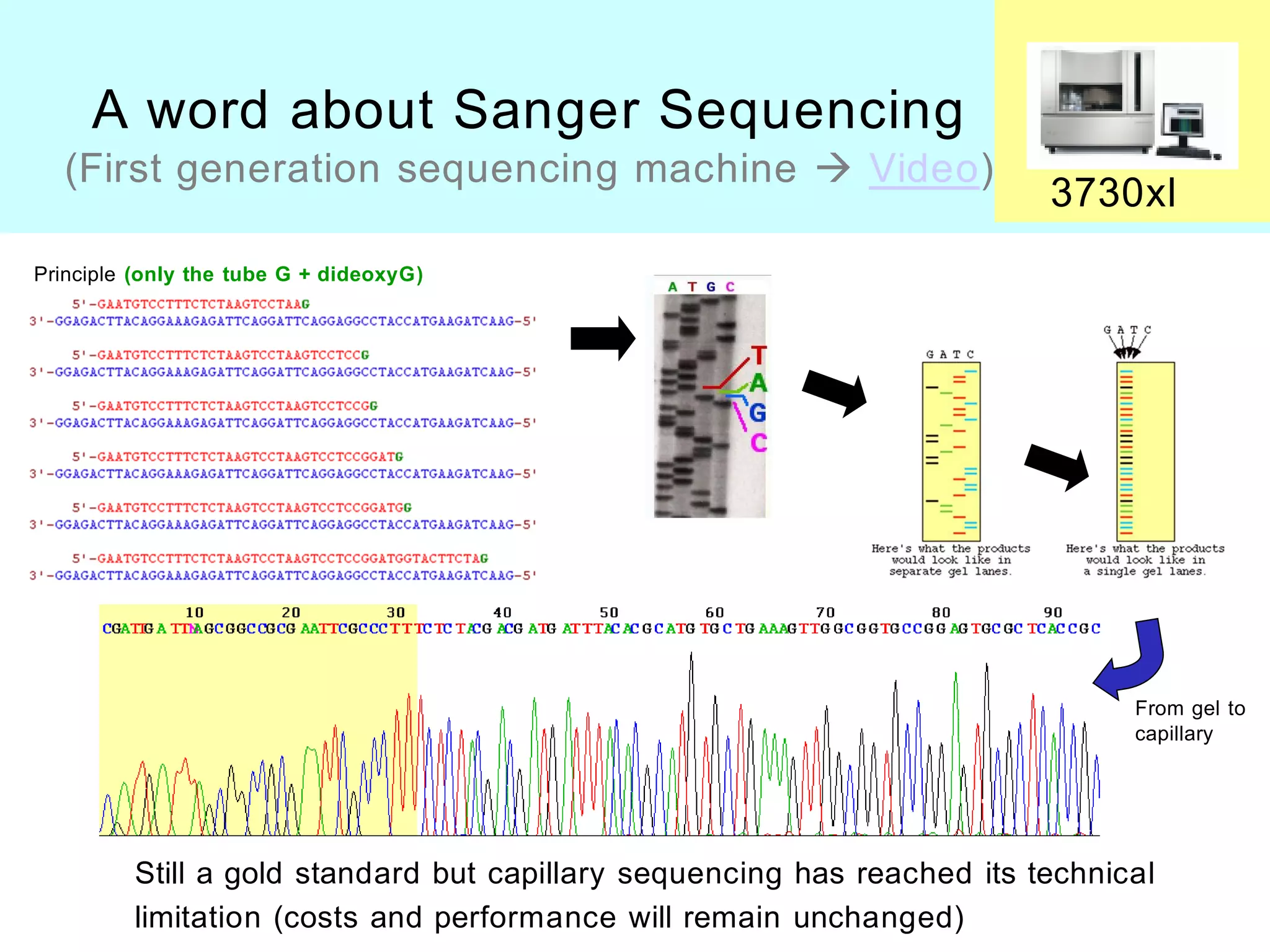
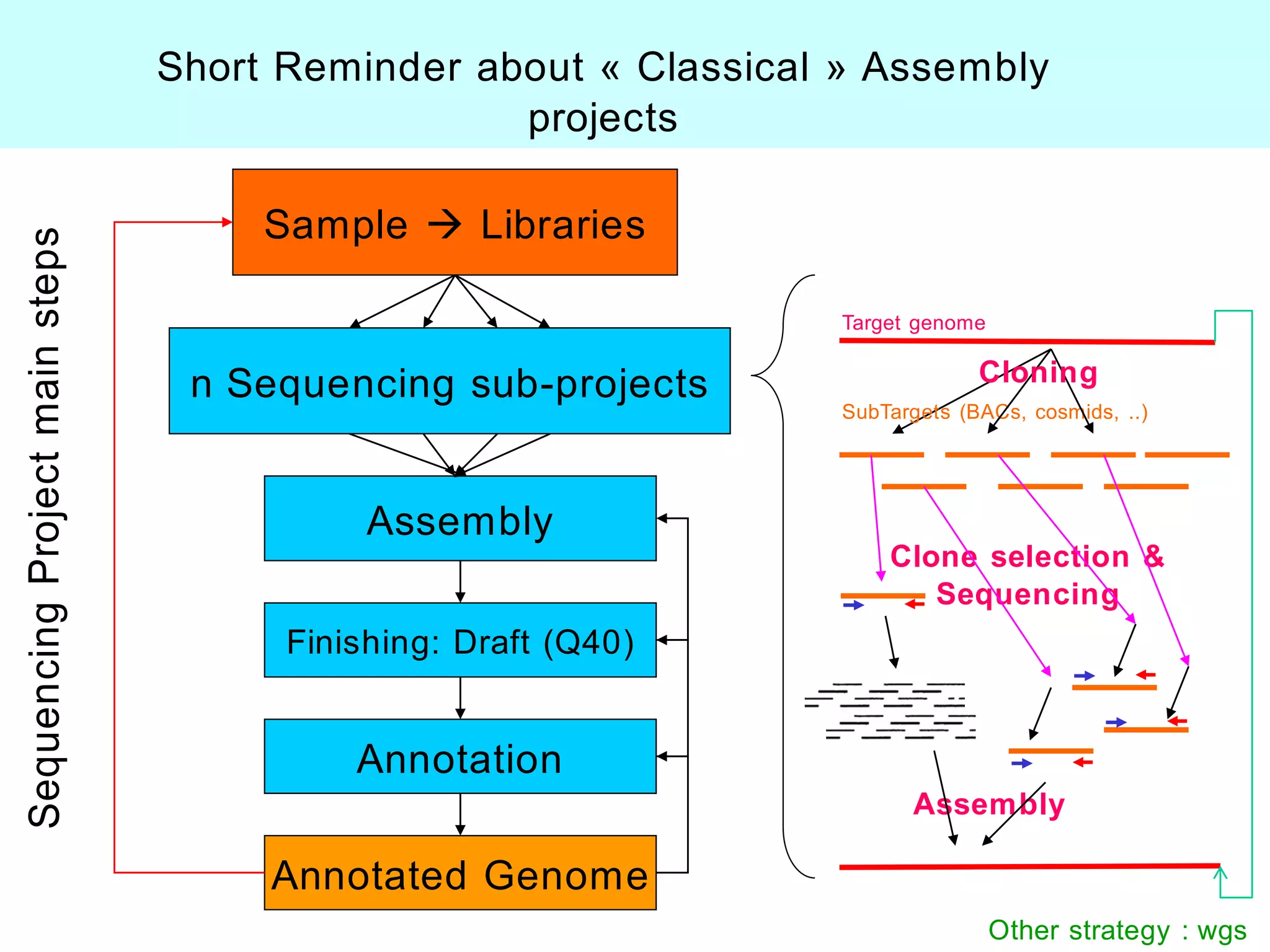
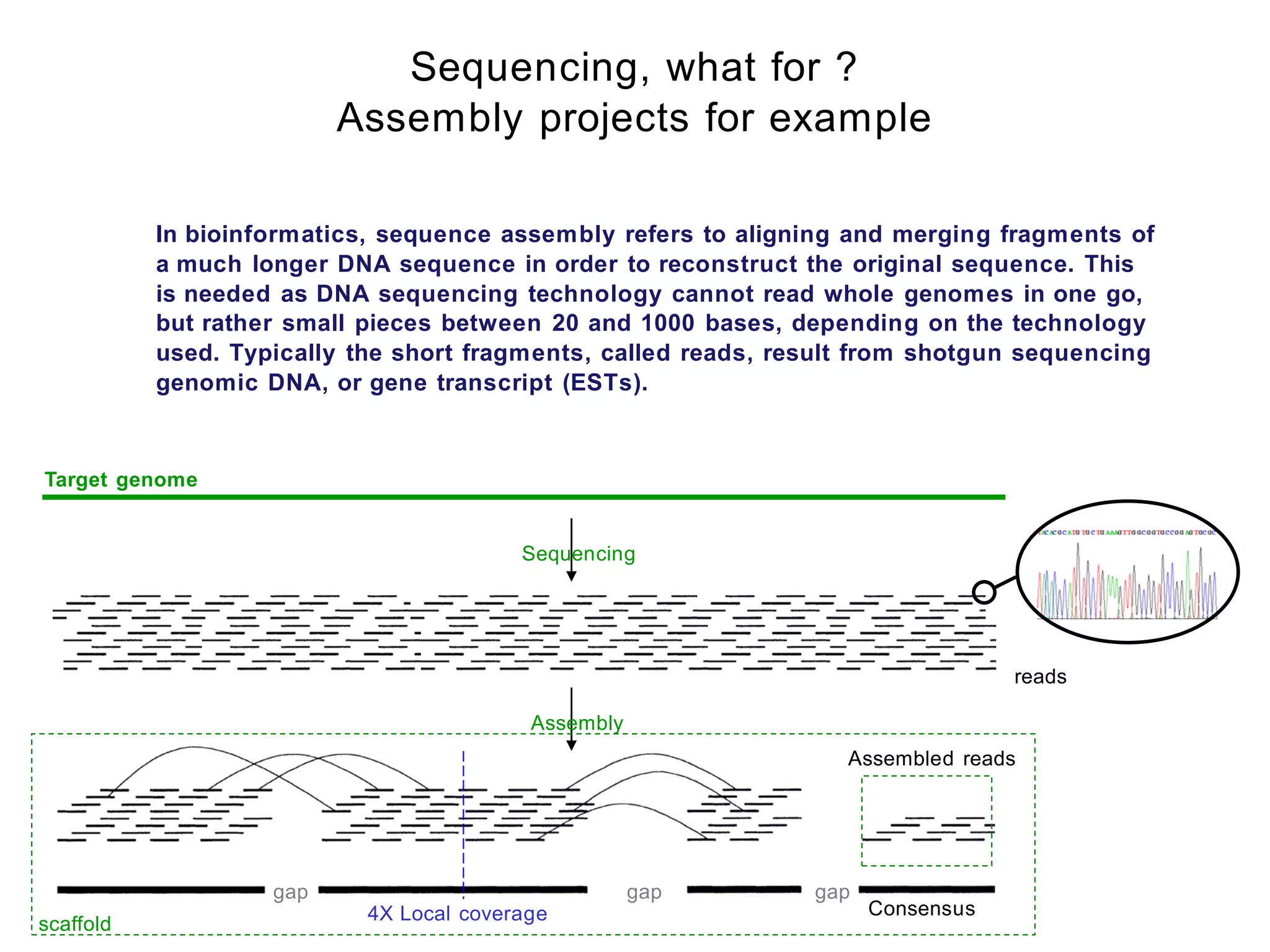
[ Reminder about Sanger Sequencing ]
• NGS Definition
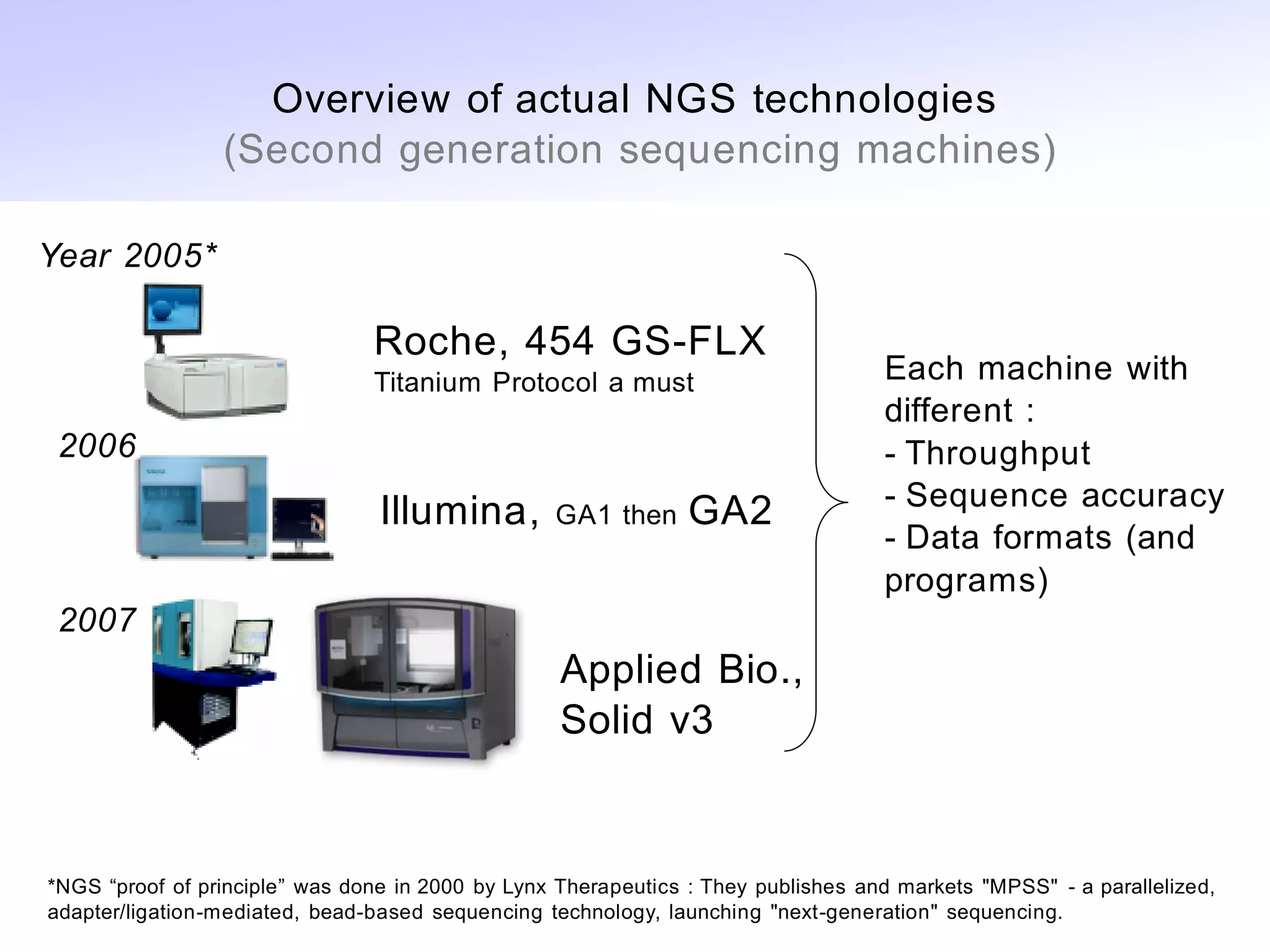
• Overview of NGS technologies
• NGS Applications & examples
• Conclusion
NOT discussed here : Sequence accuracy, assembly and sampling ; NGS
data Analysis & BioInformatics tools](https://image.slidesharecdn.com/ngsintrov6public-110320062557-phpapp01/75/Ngs-intro_v6_public-2-2048.jpg)