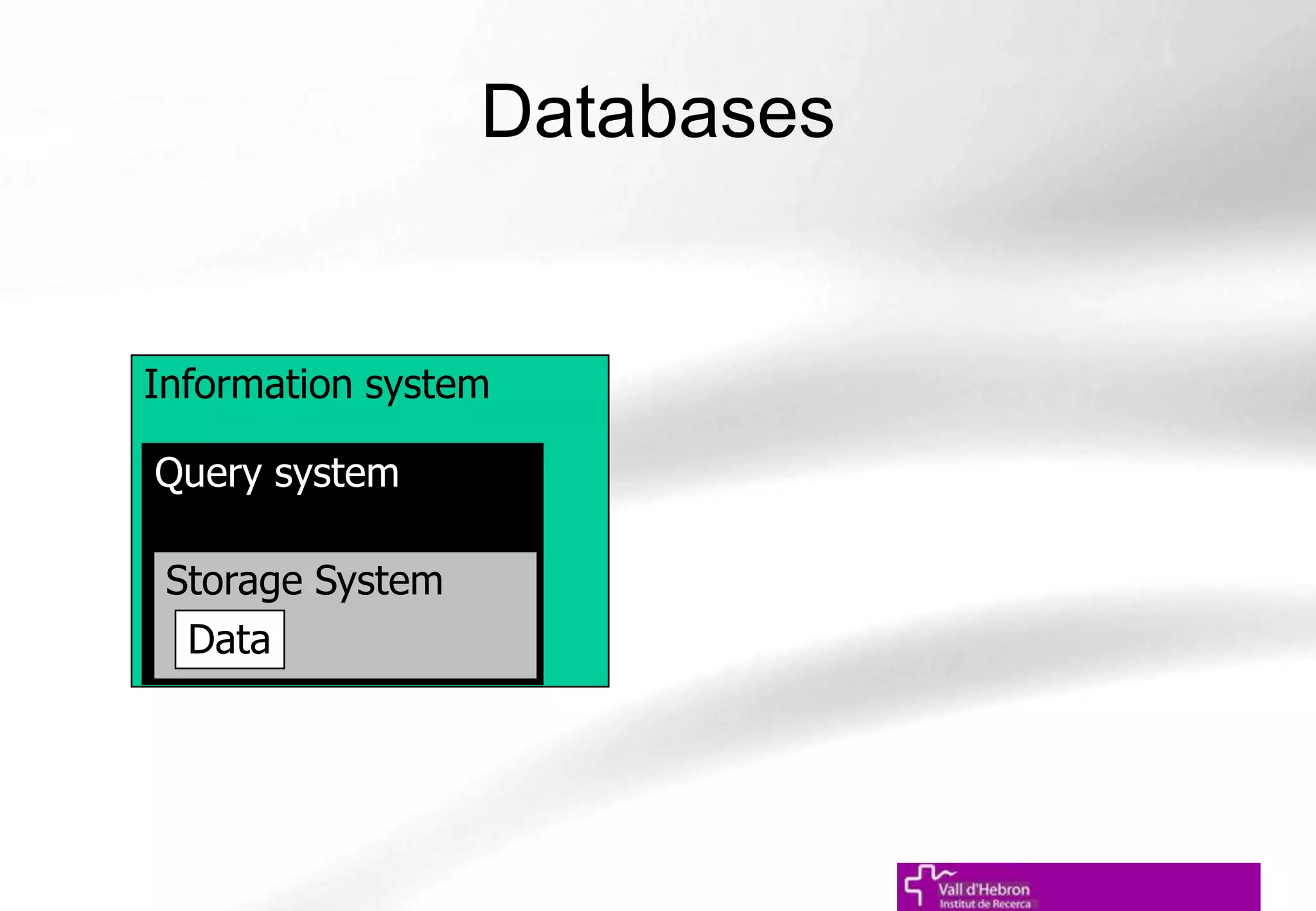
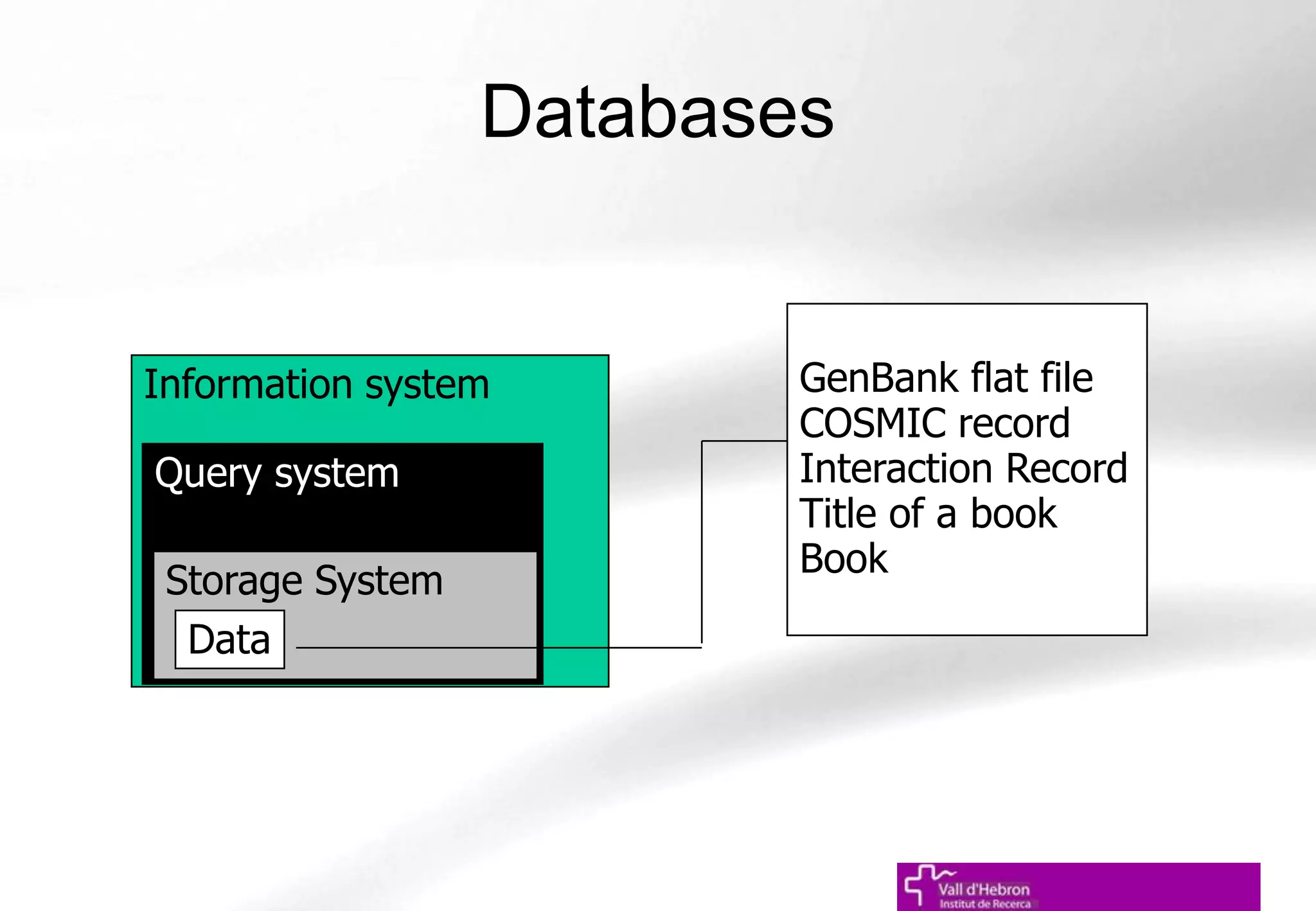
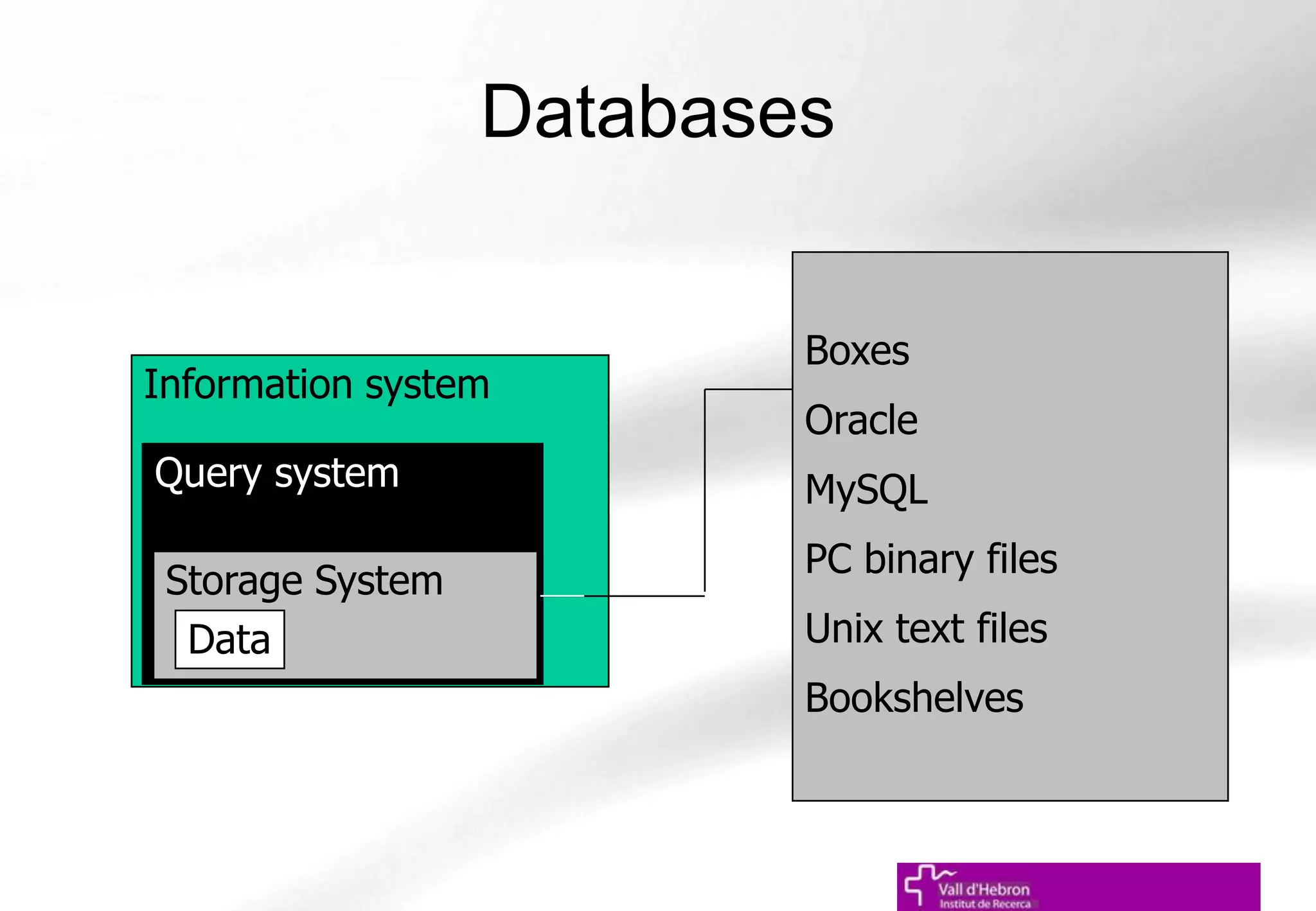
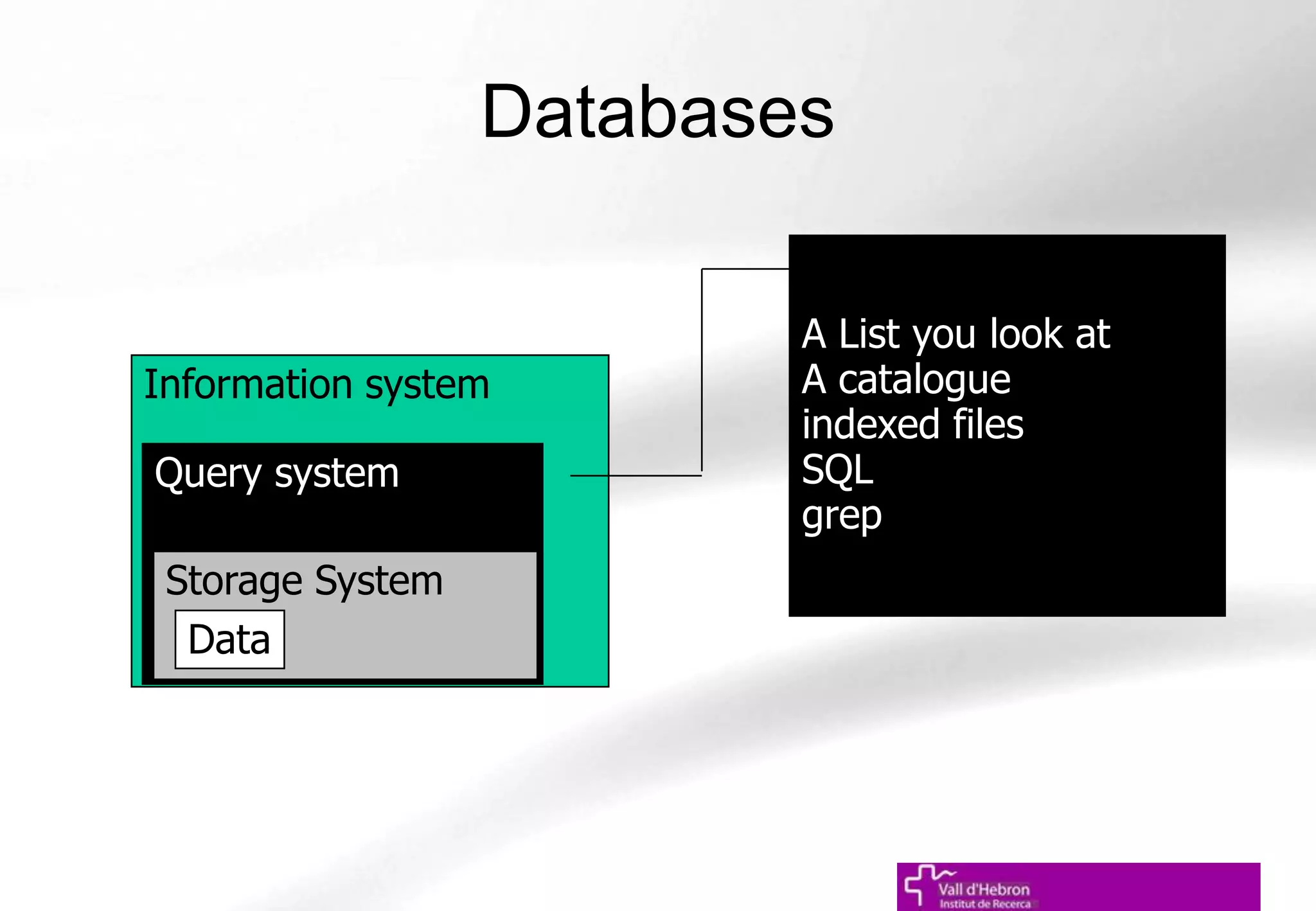
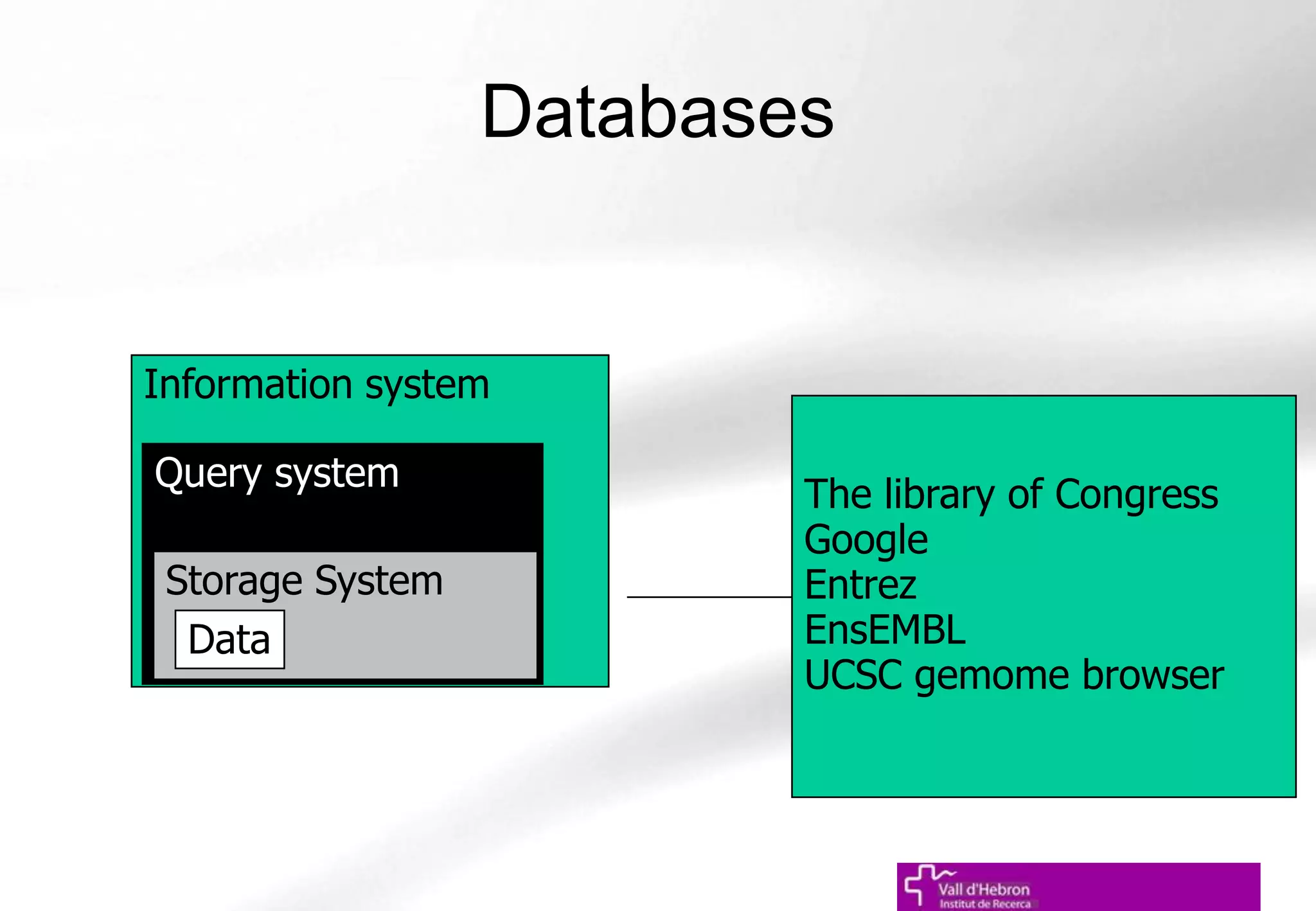
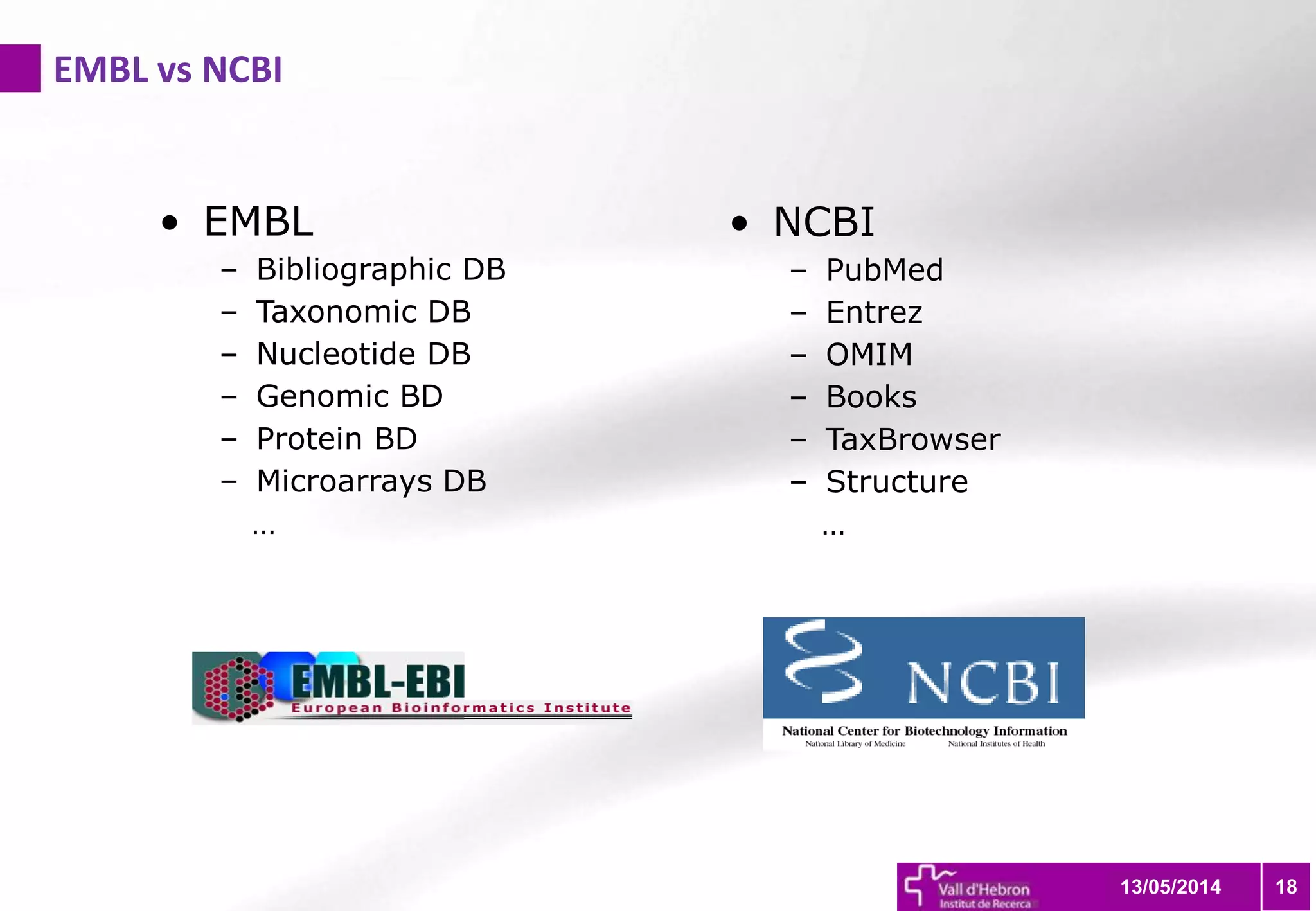
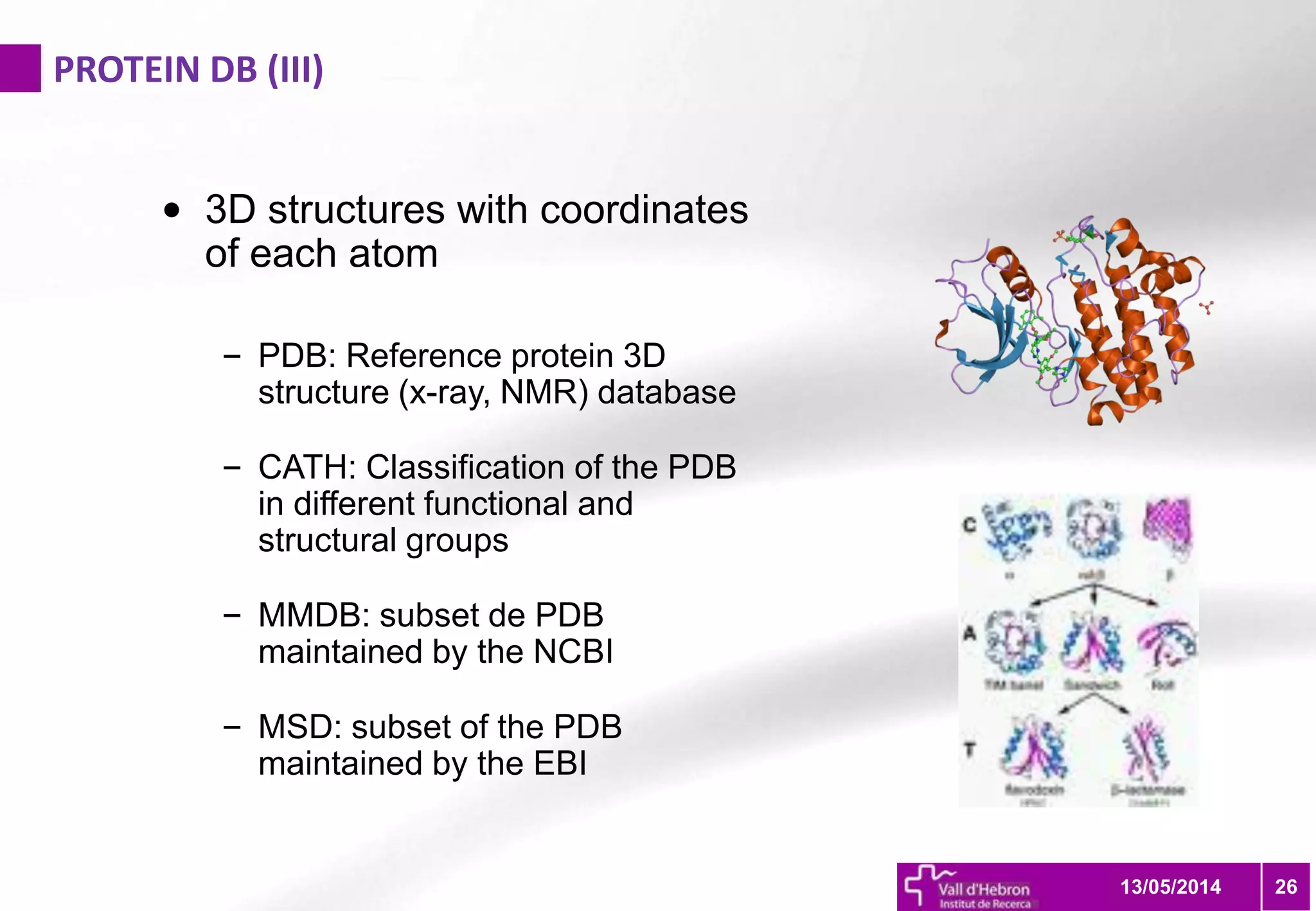
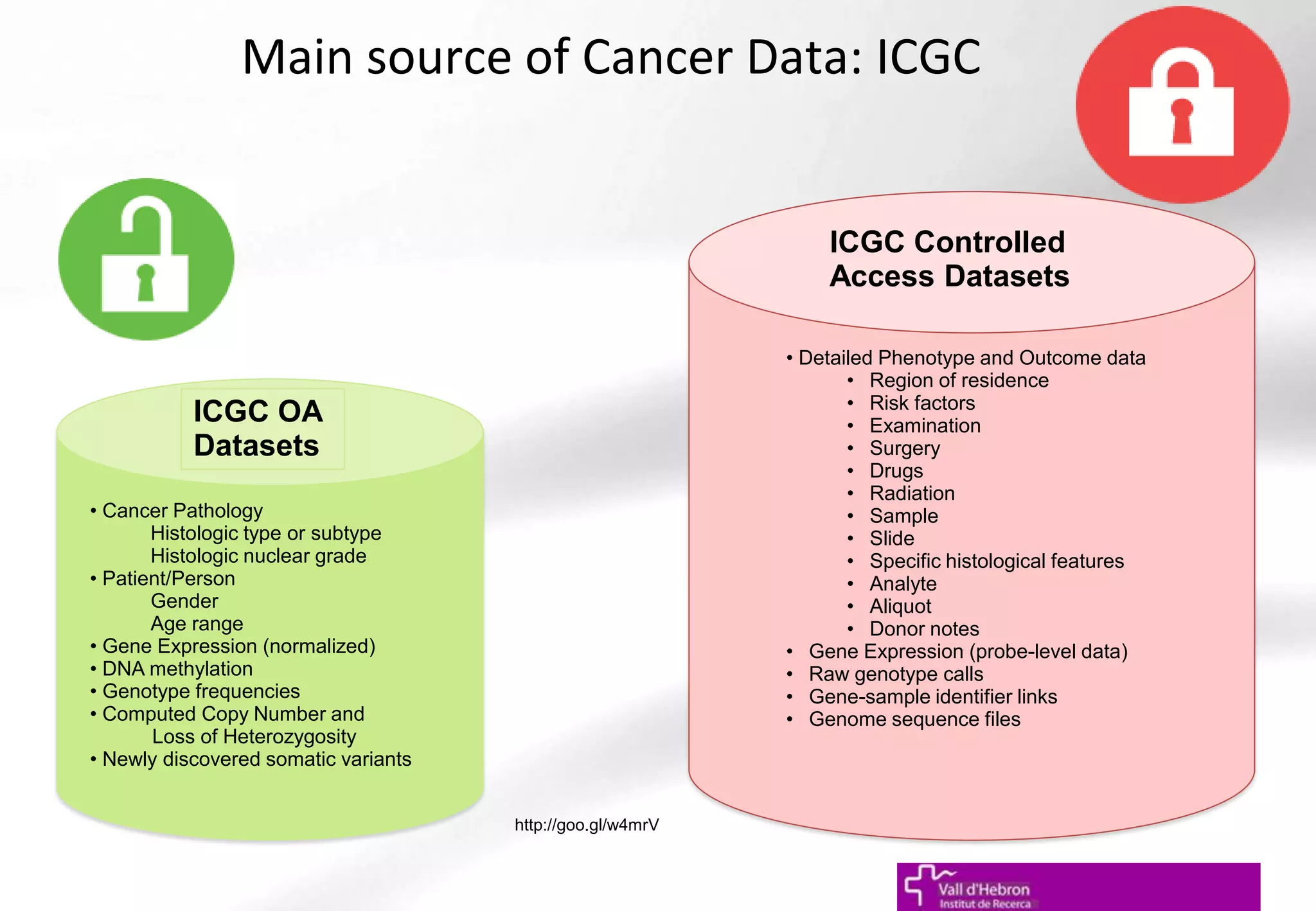
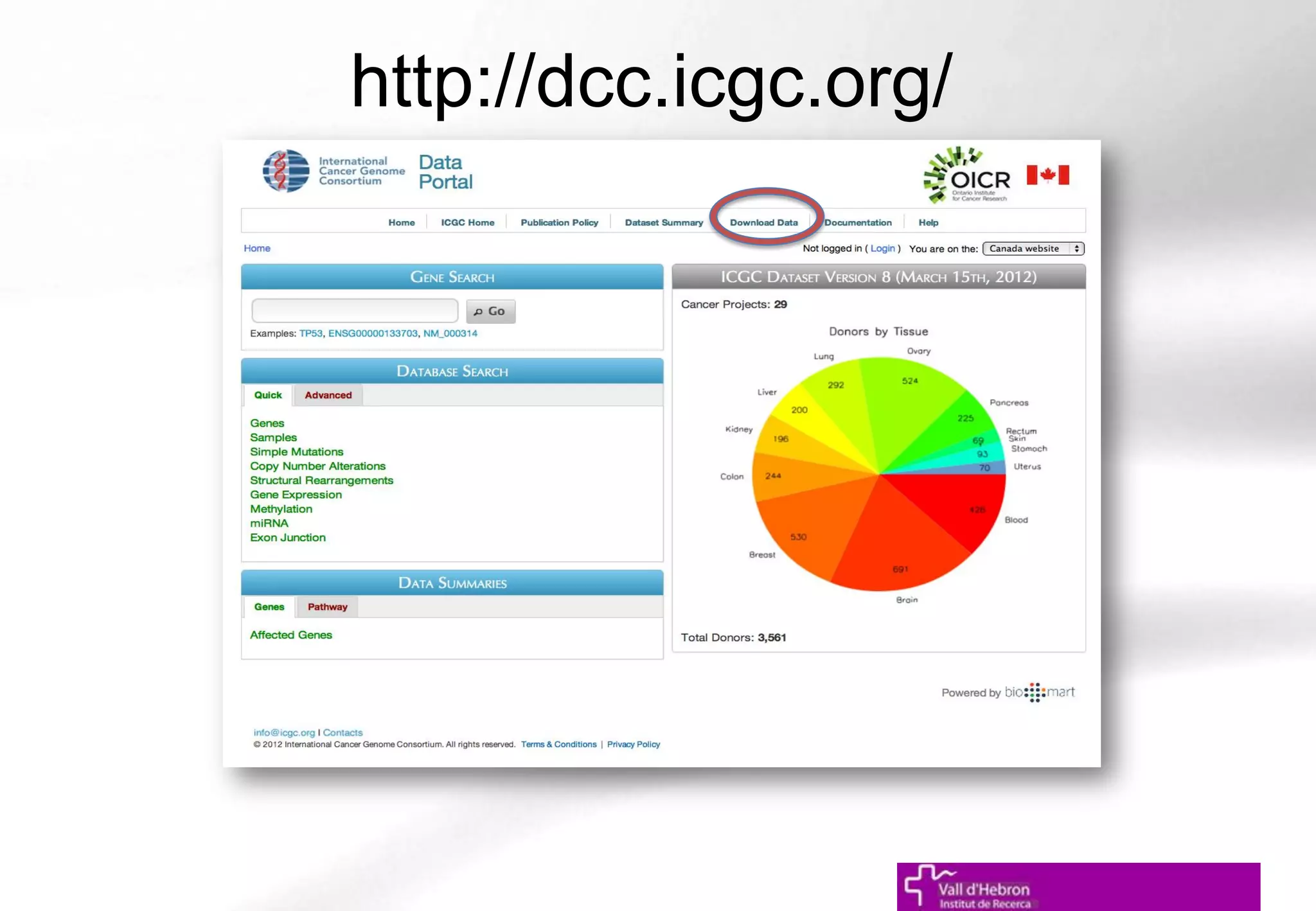
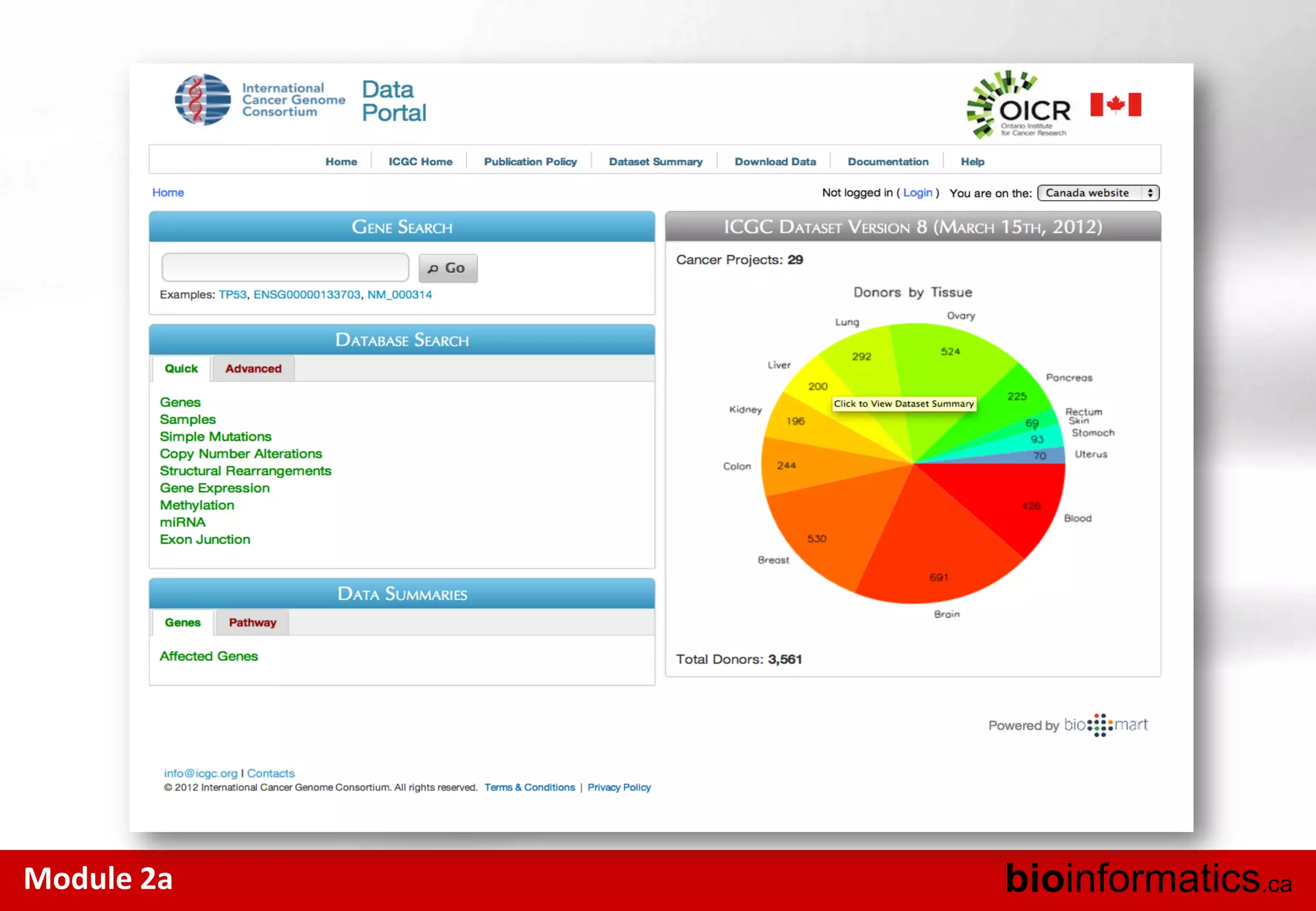
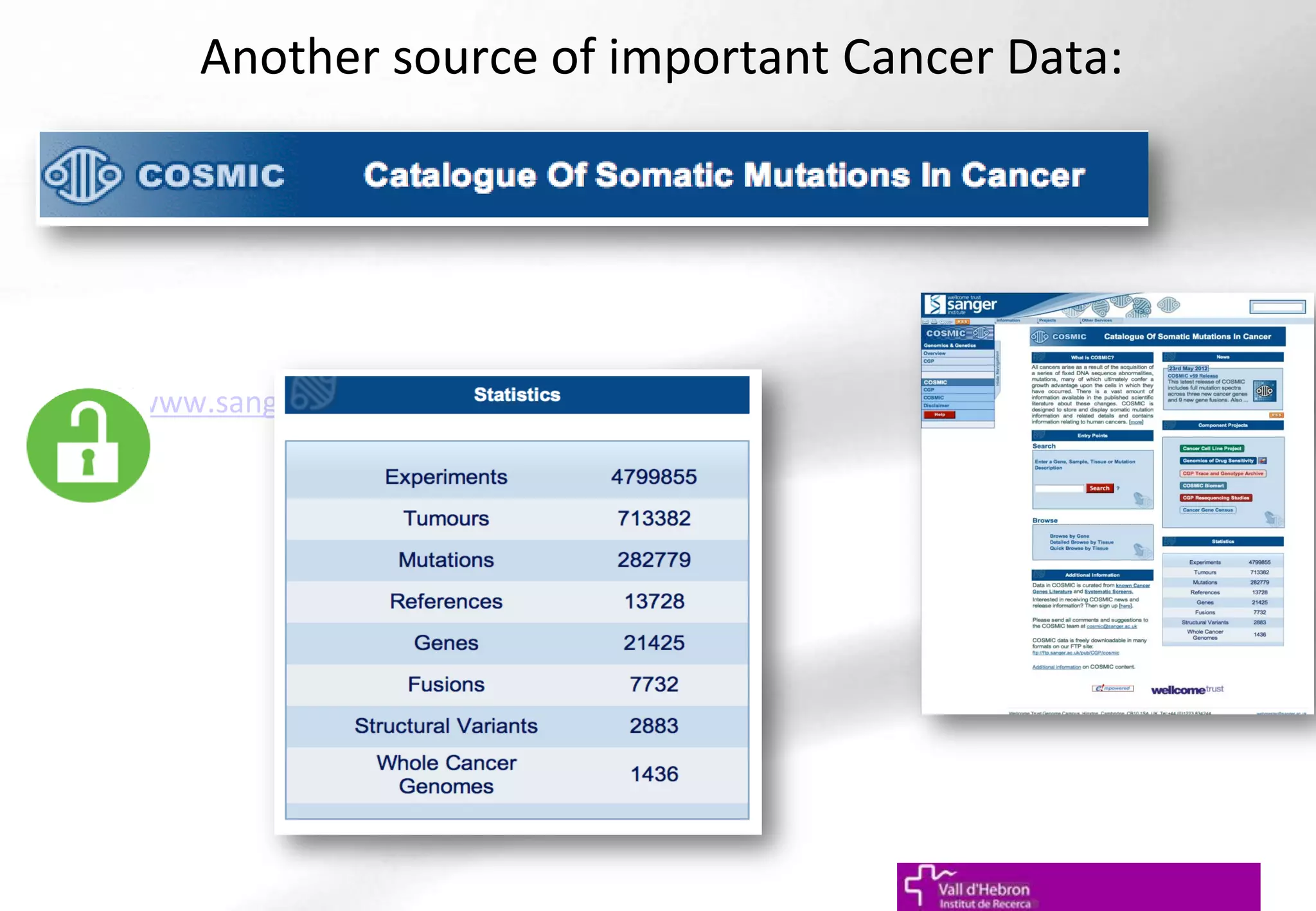
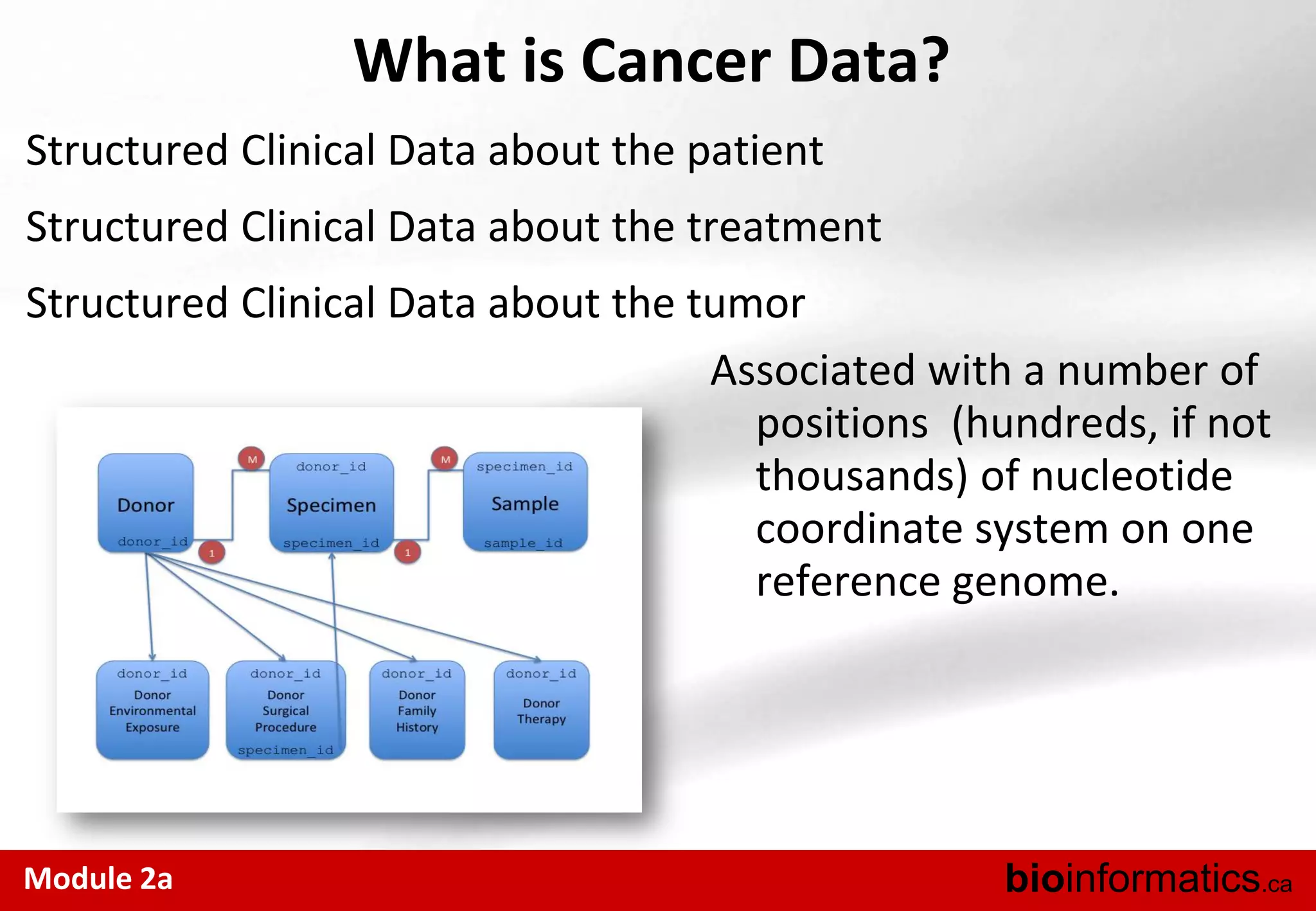
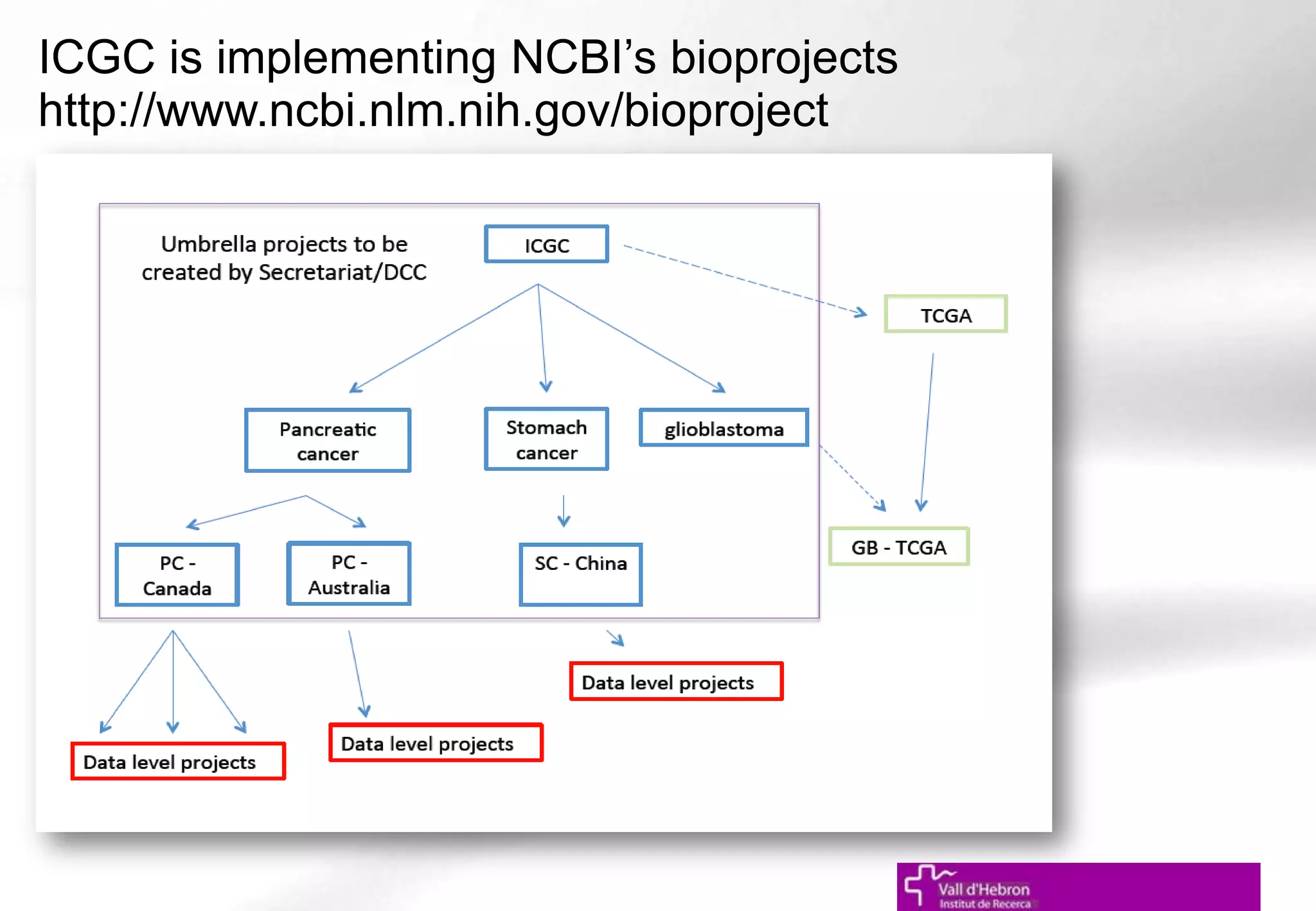
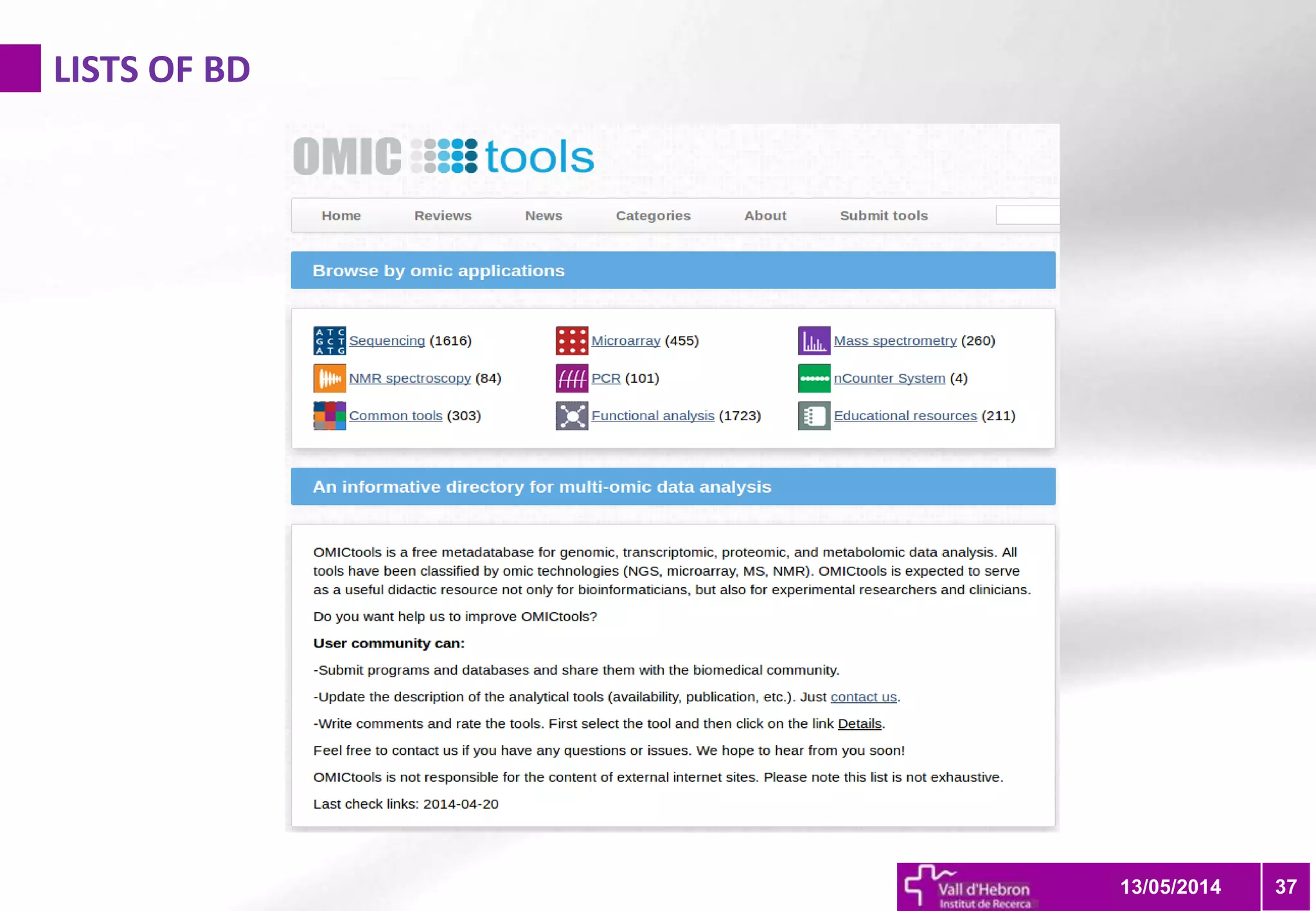
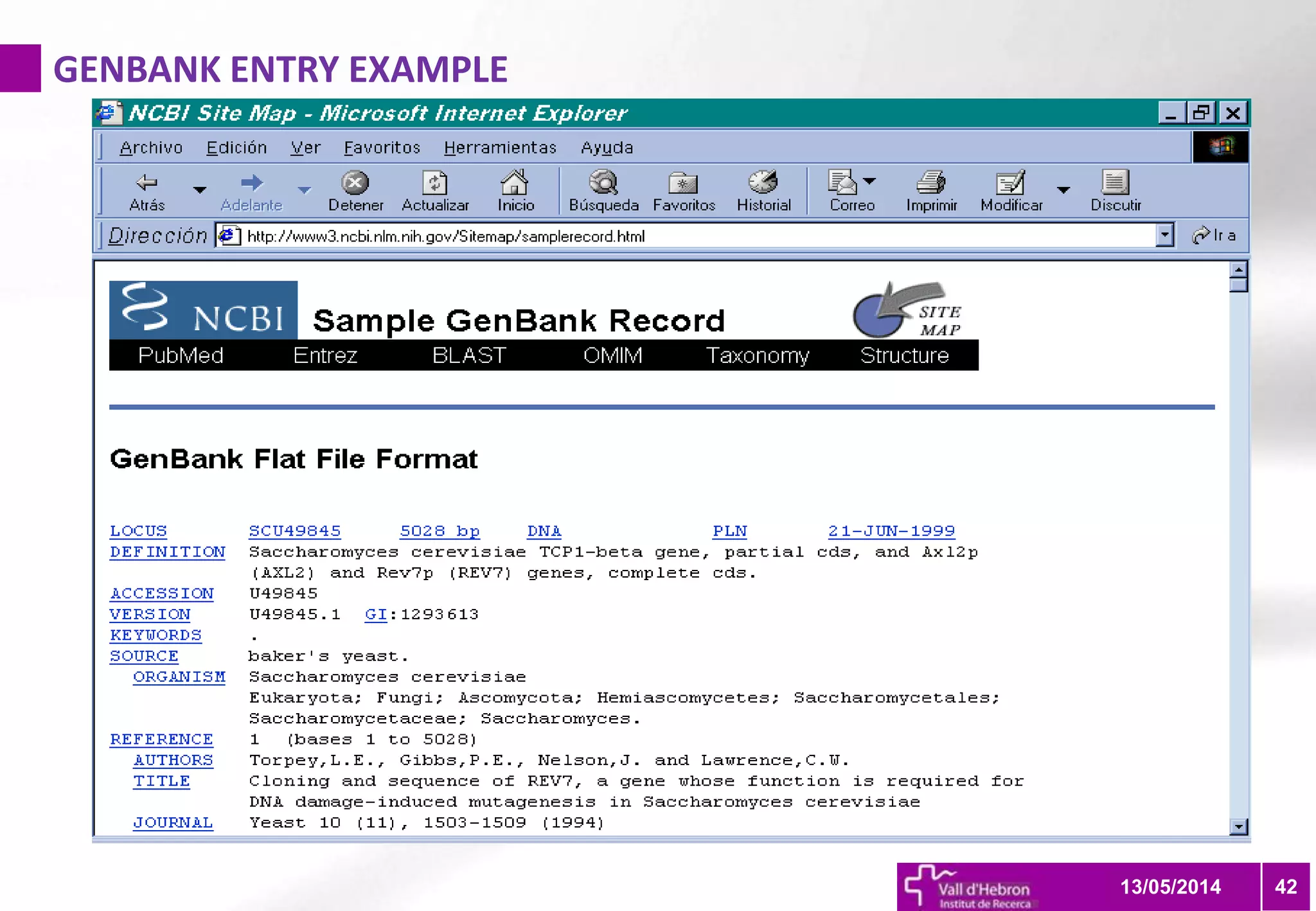
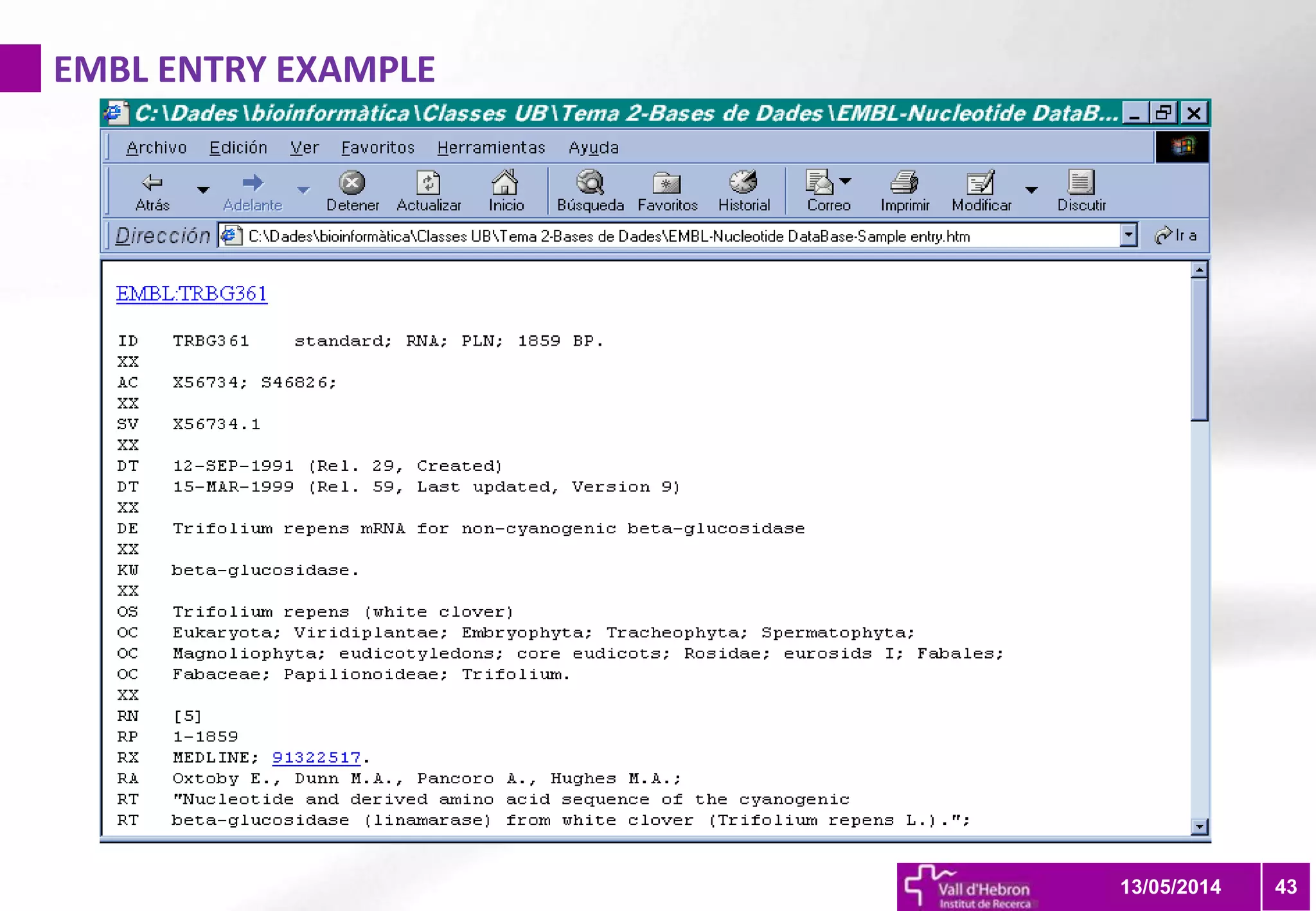
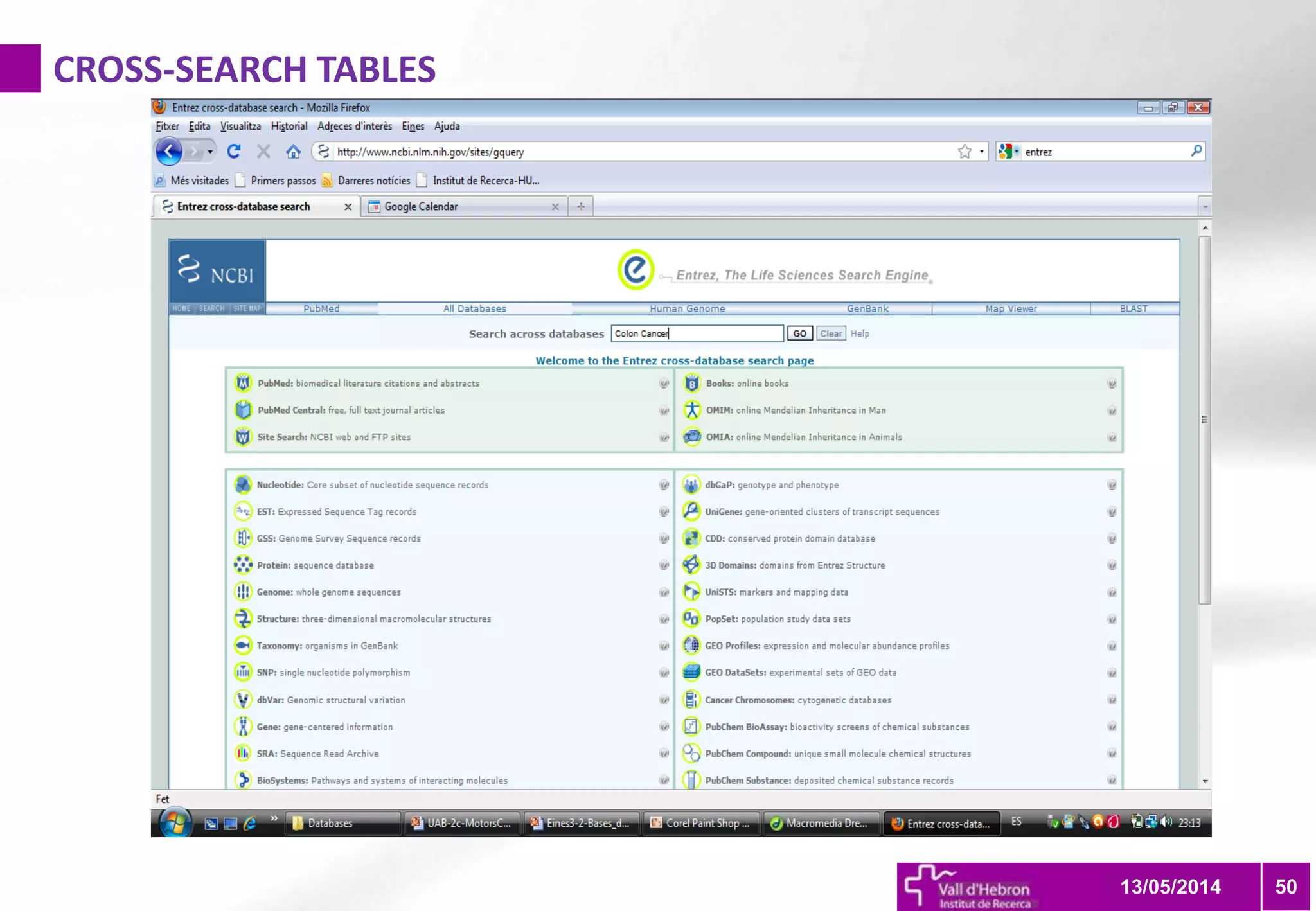
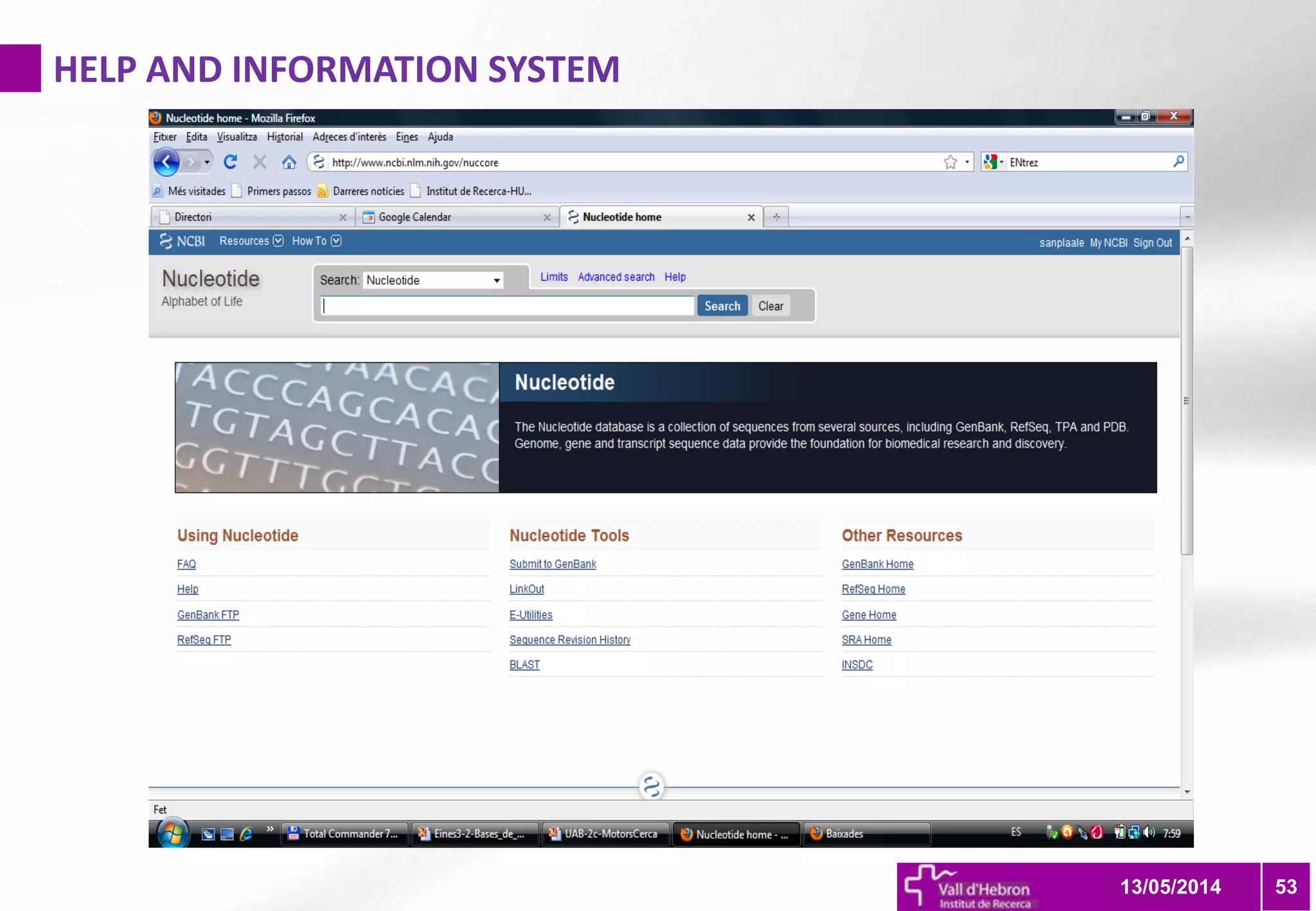
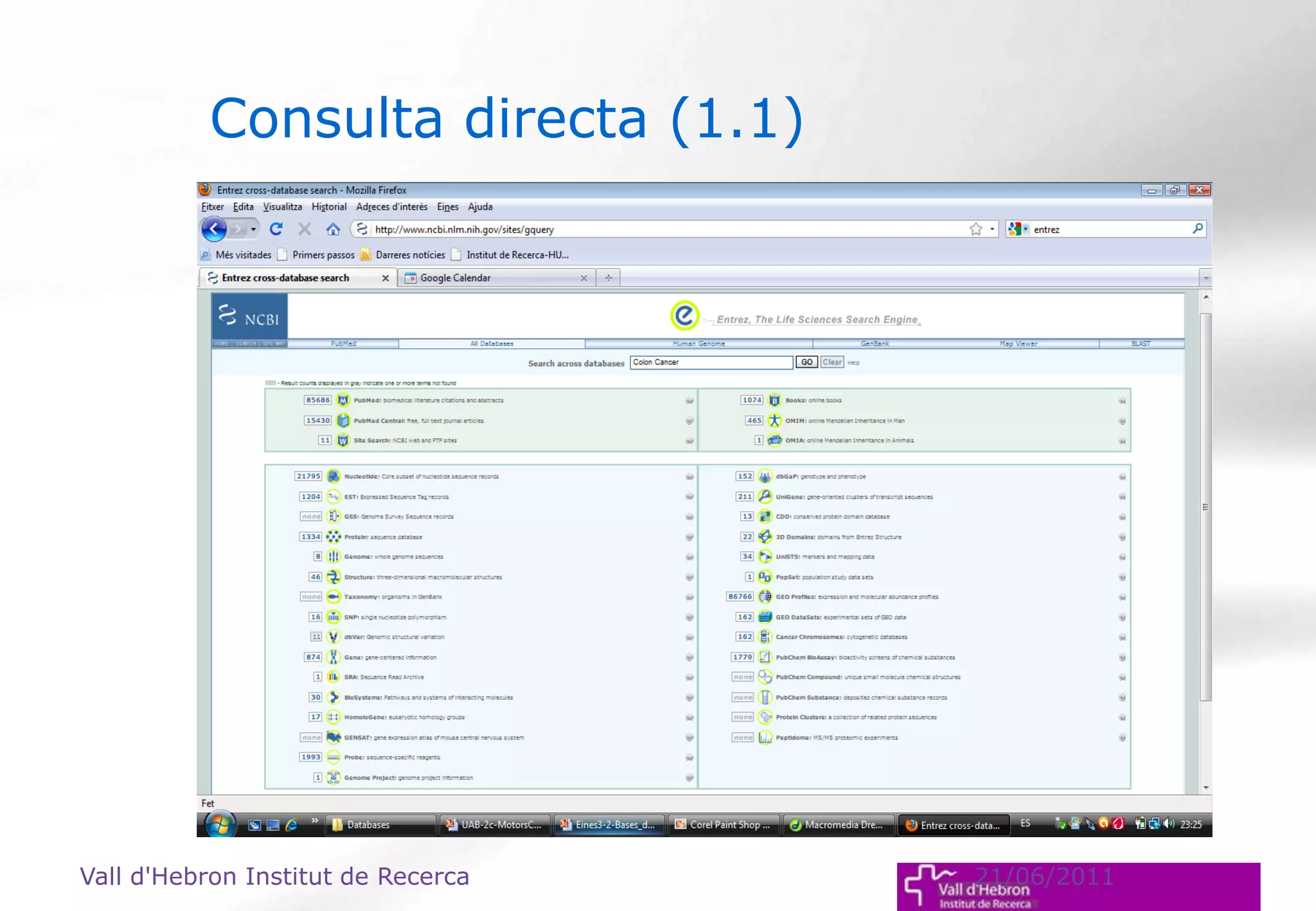
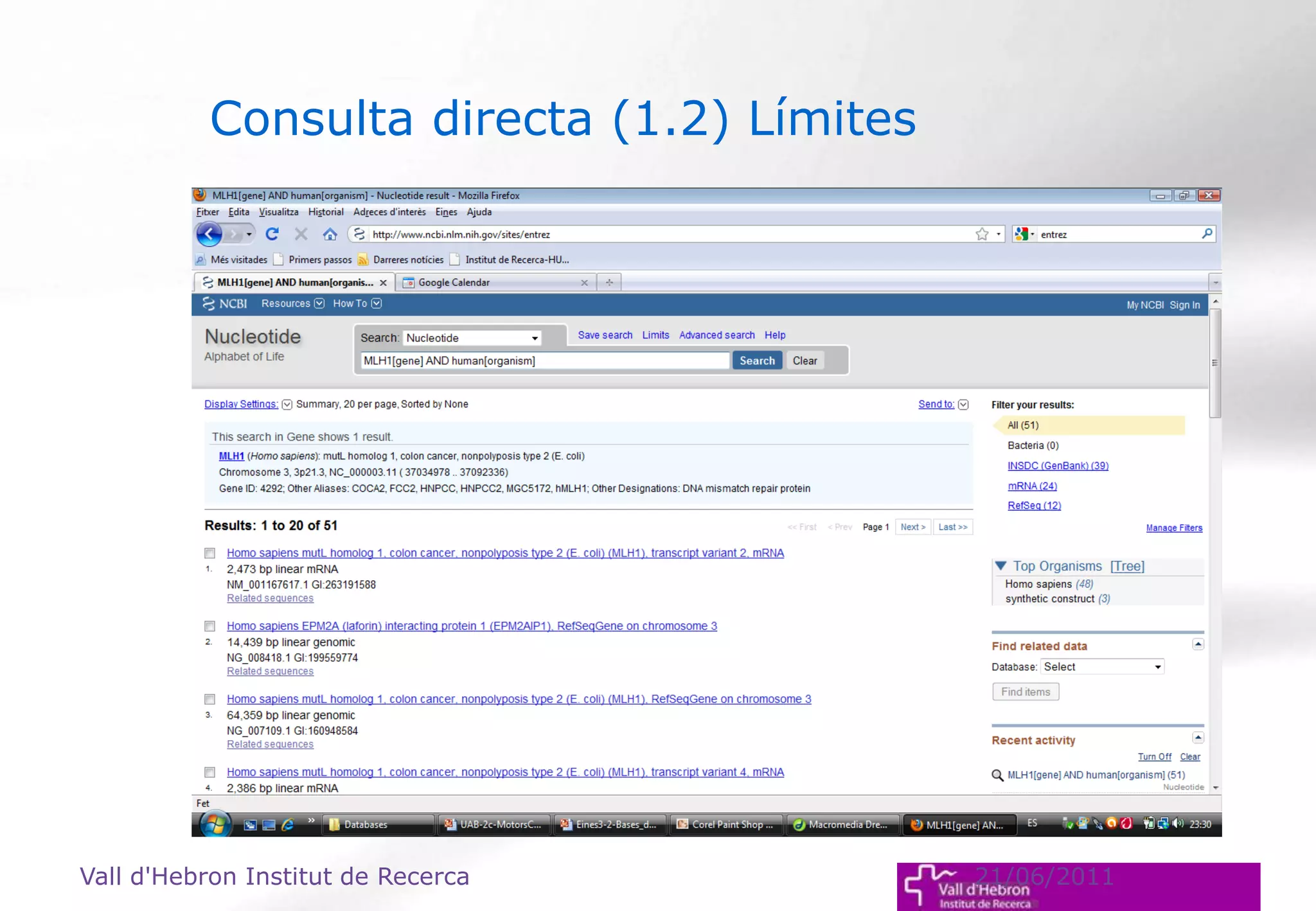
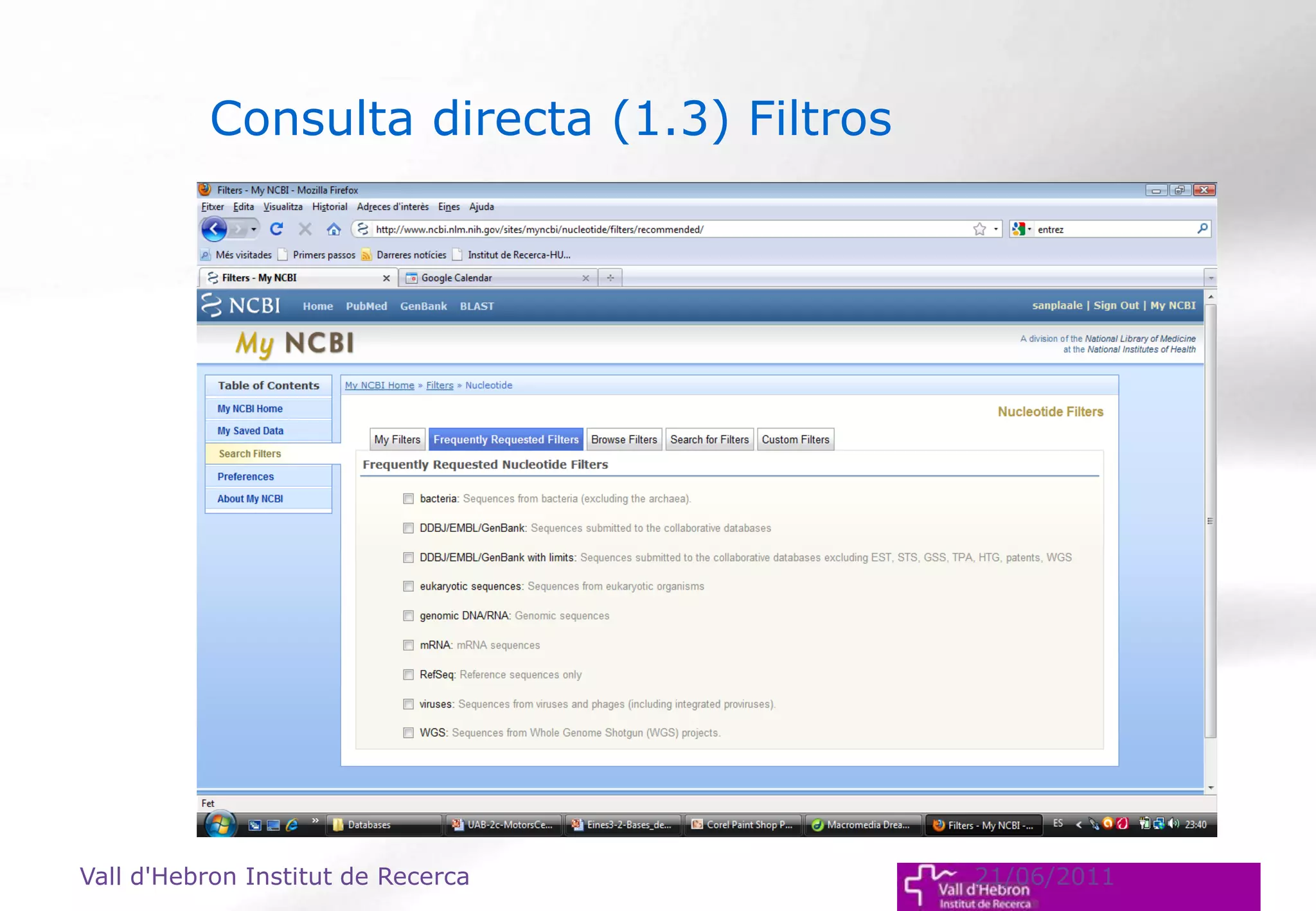
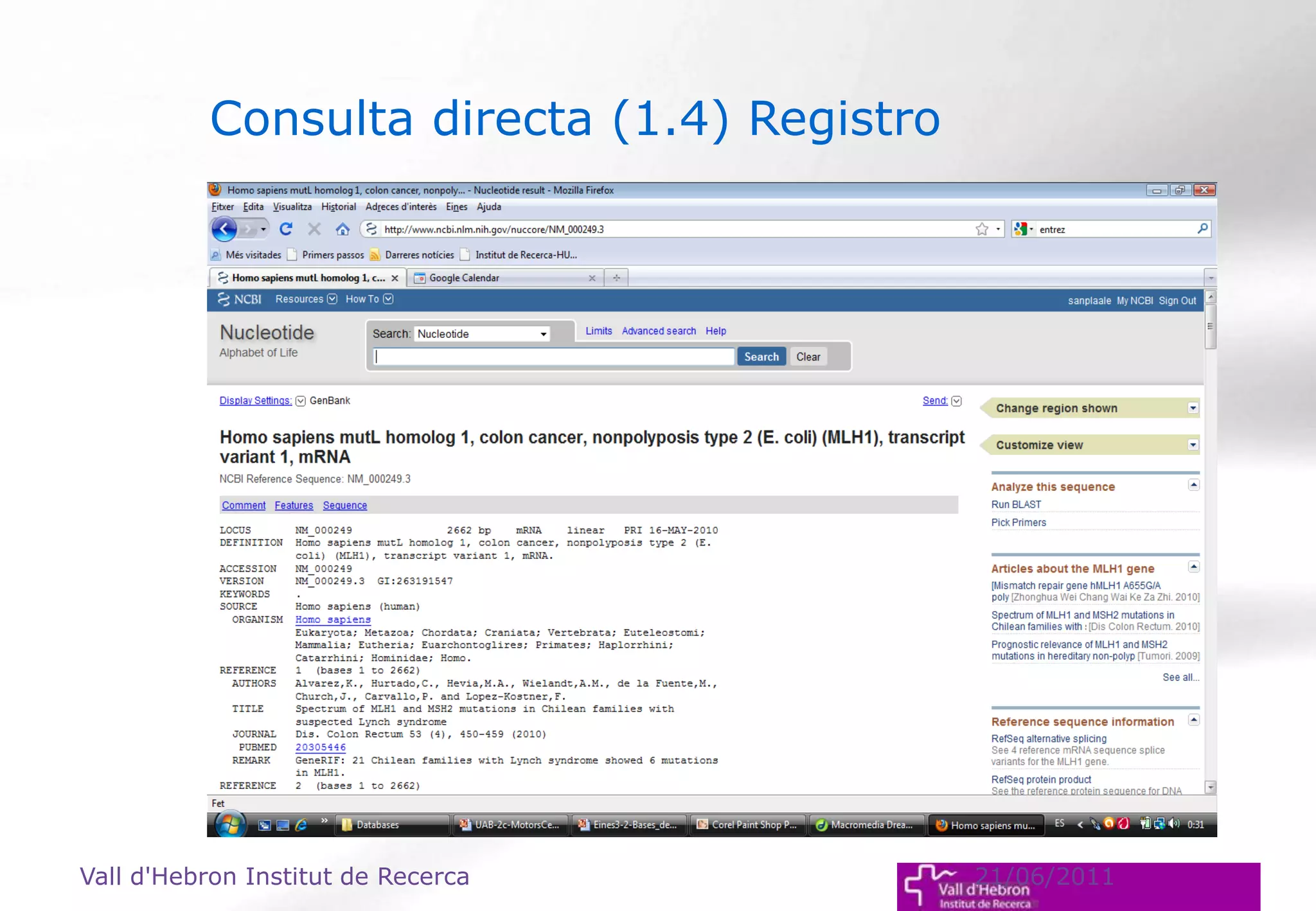
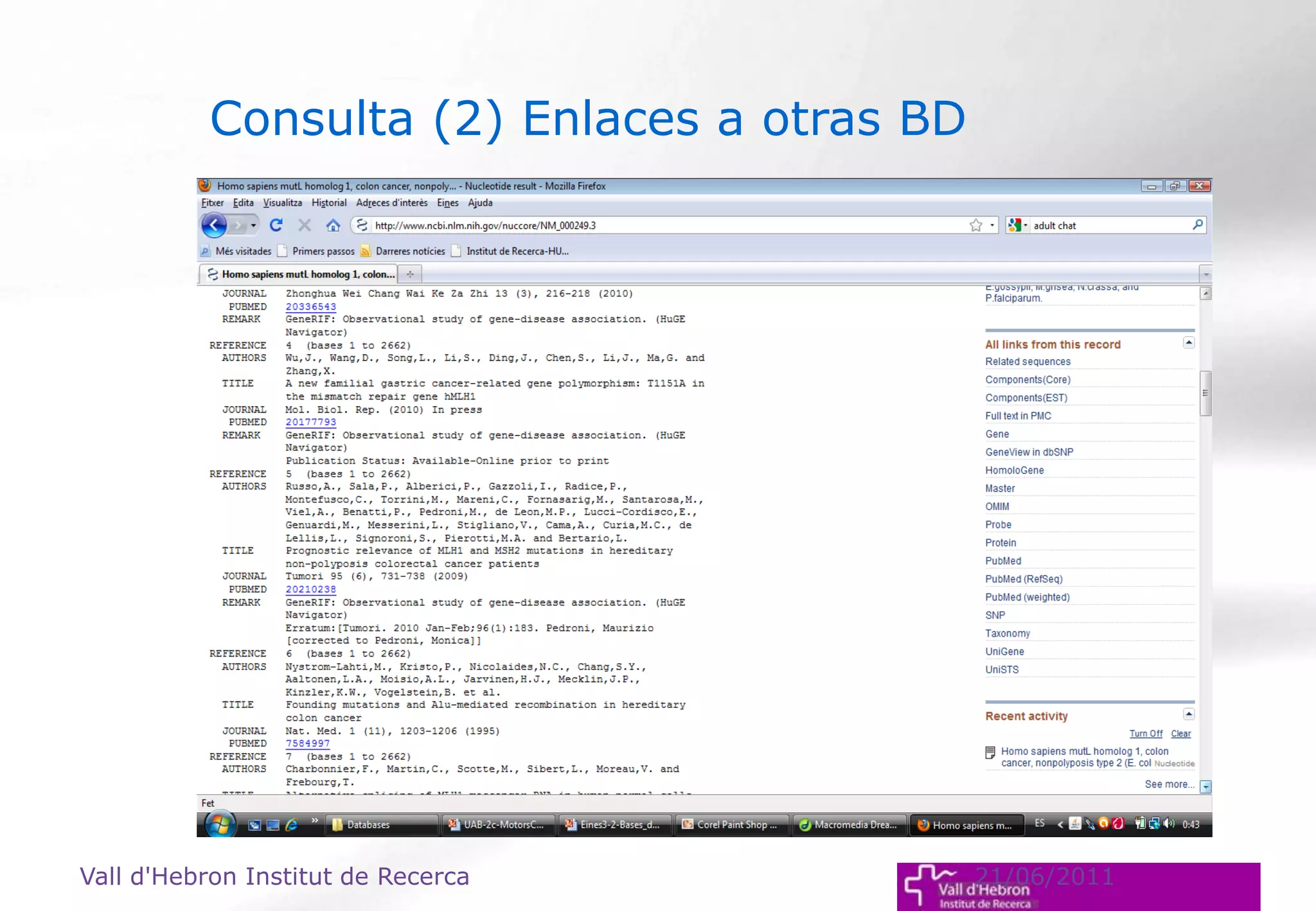
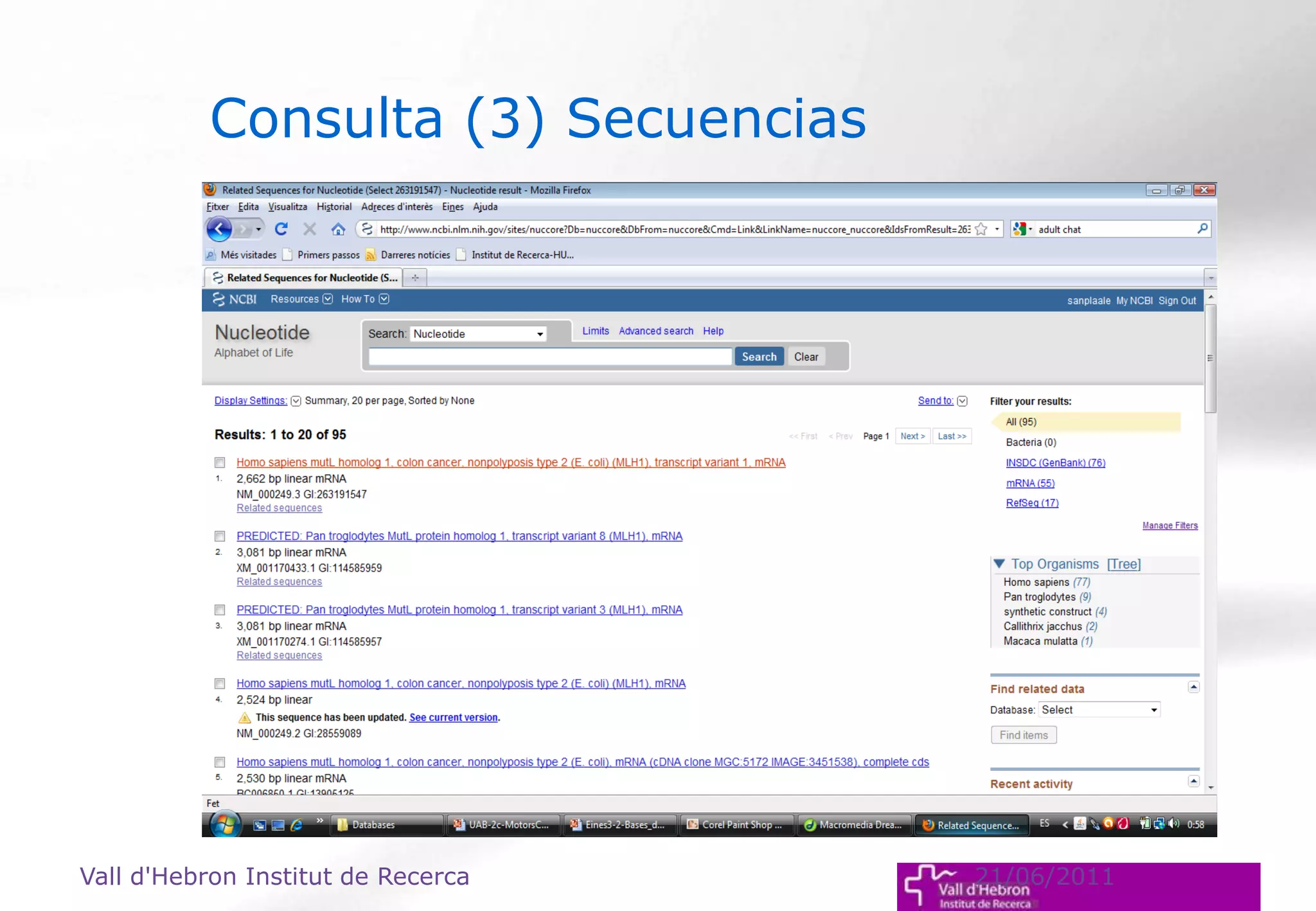
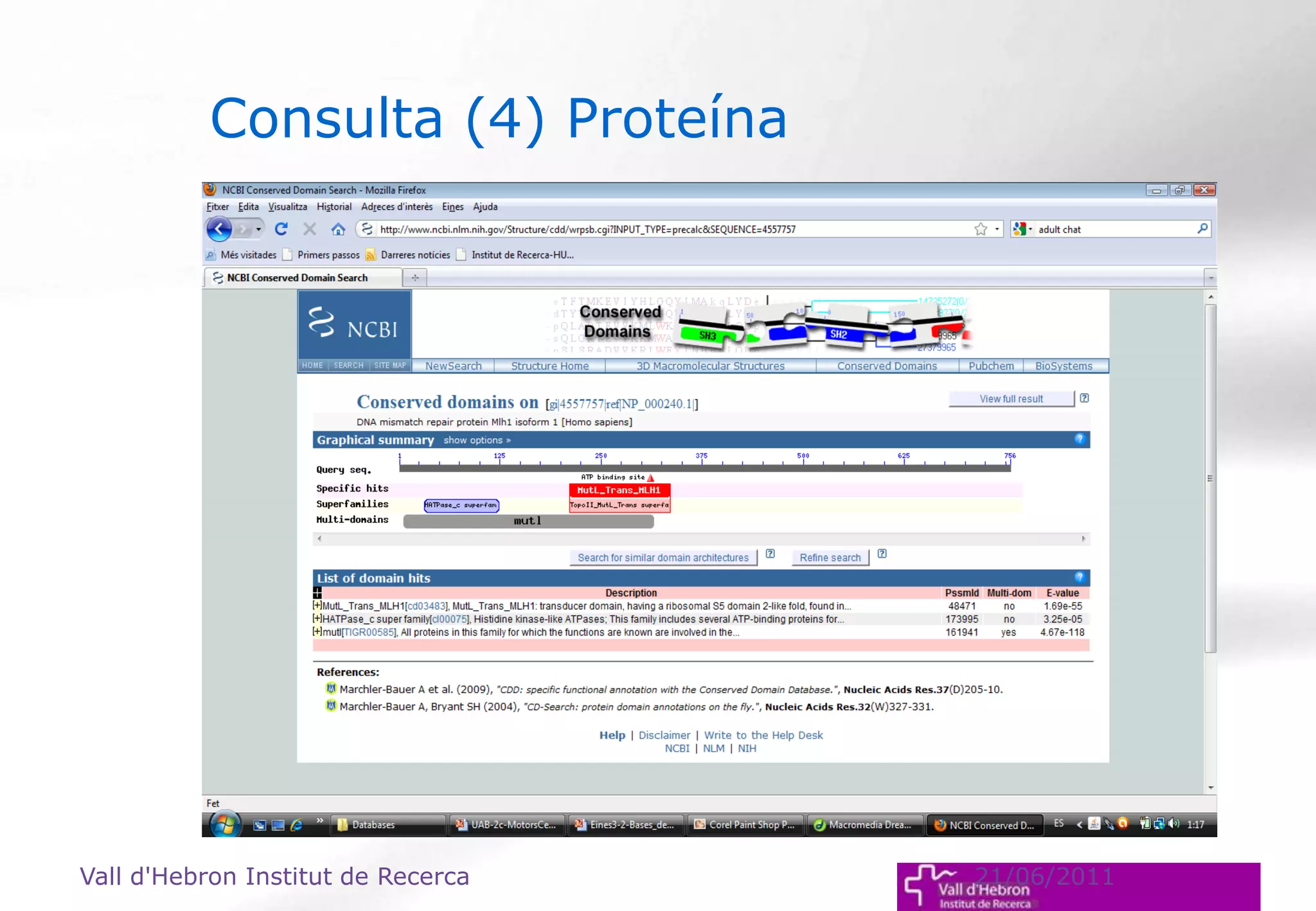
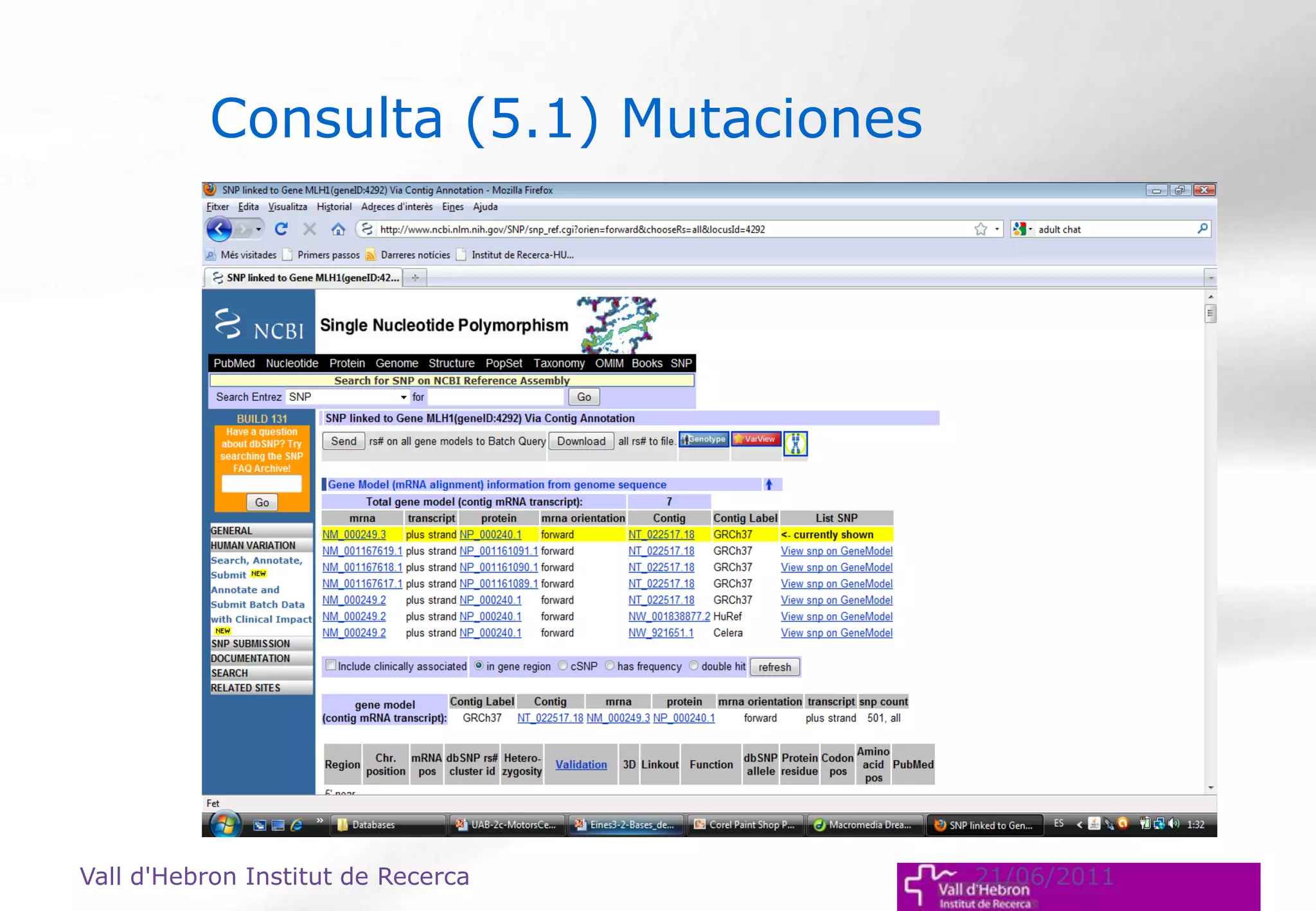
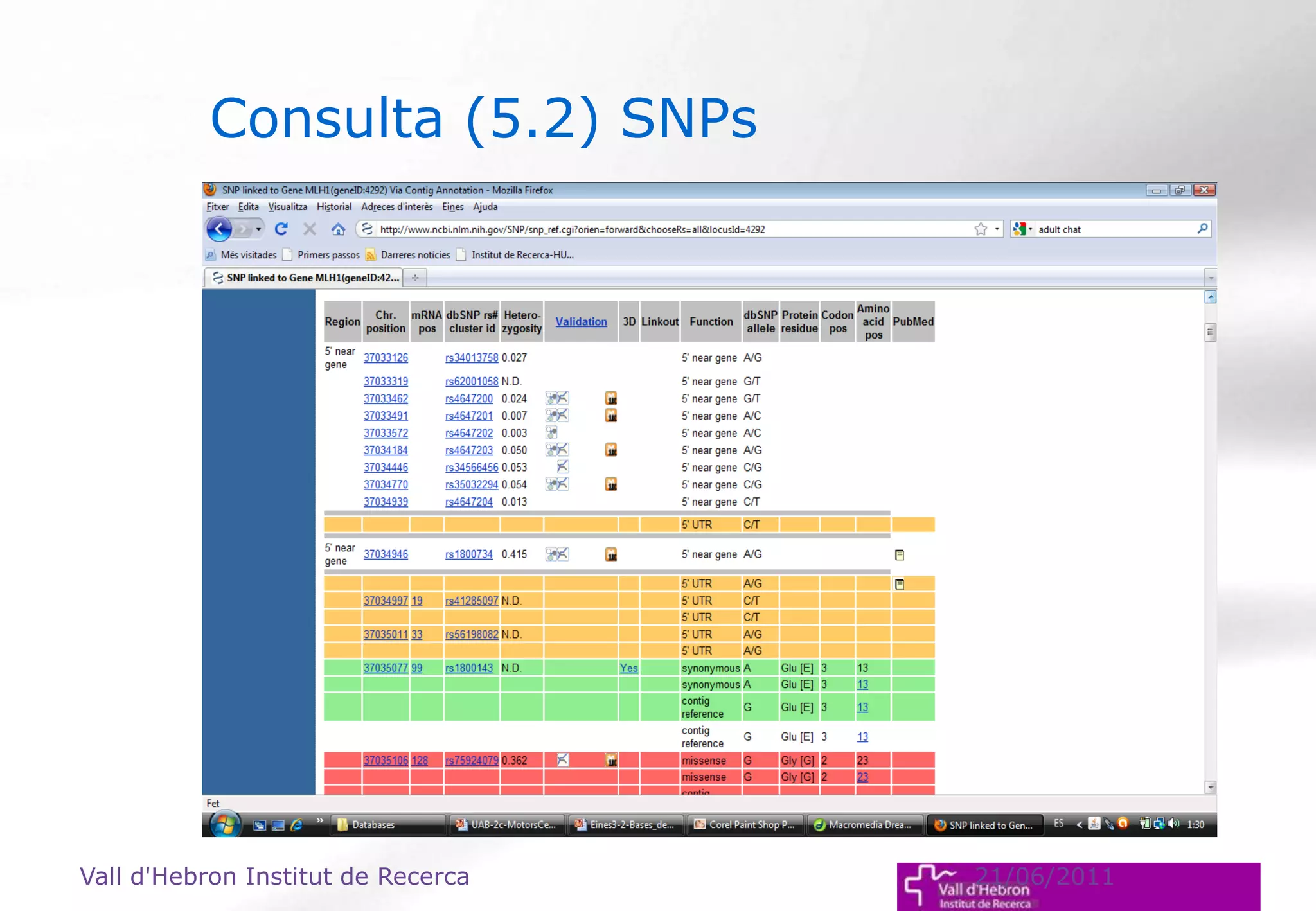
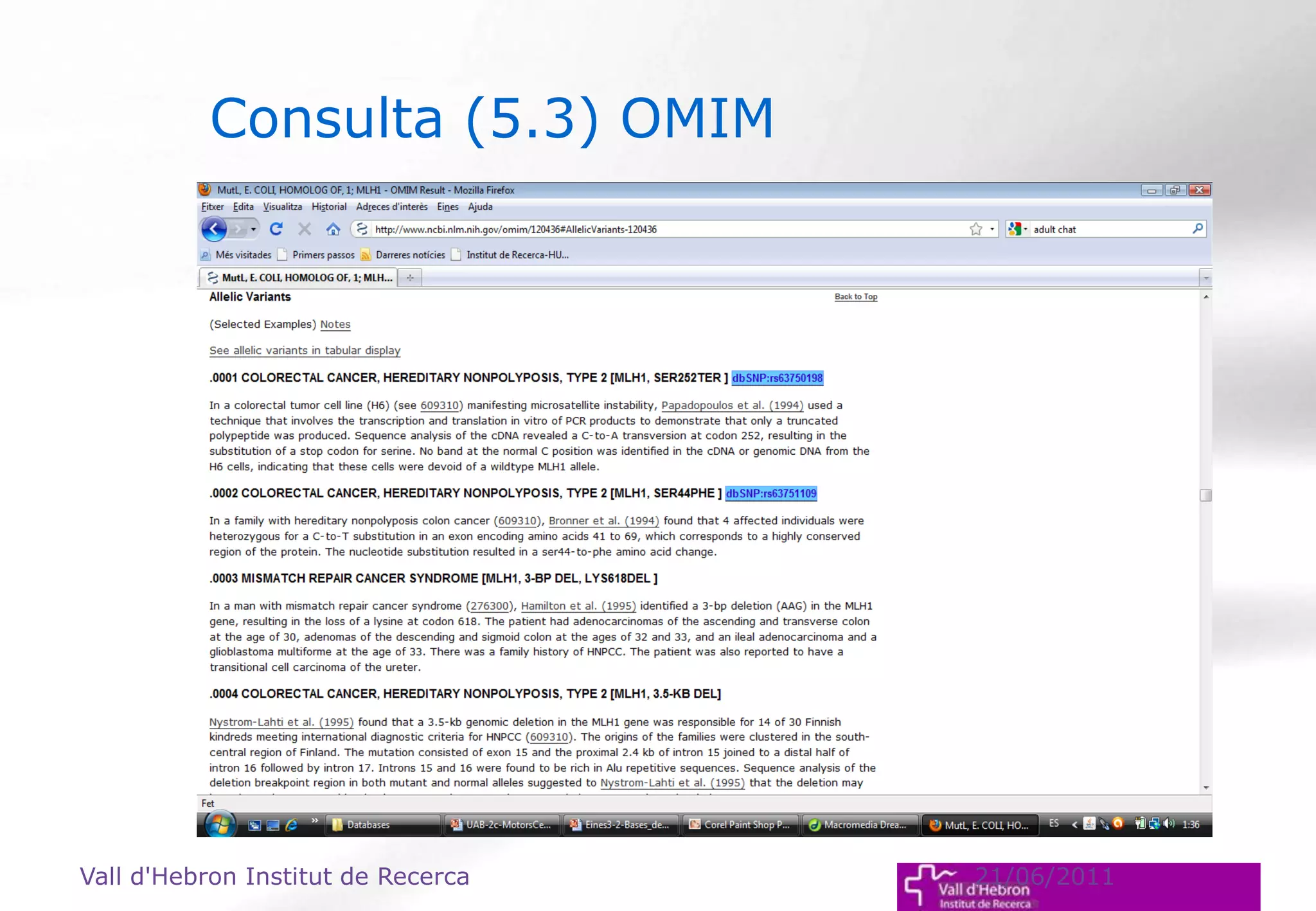
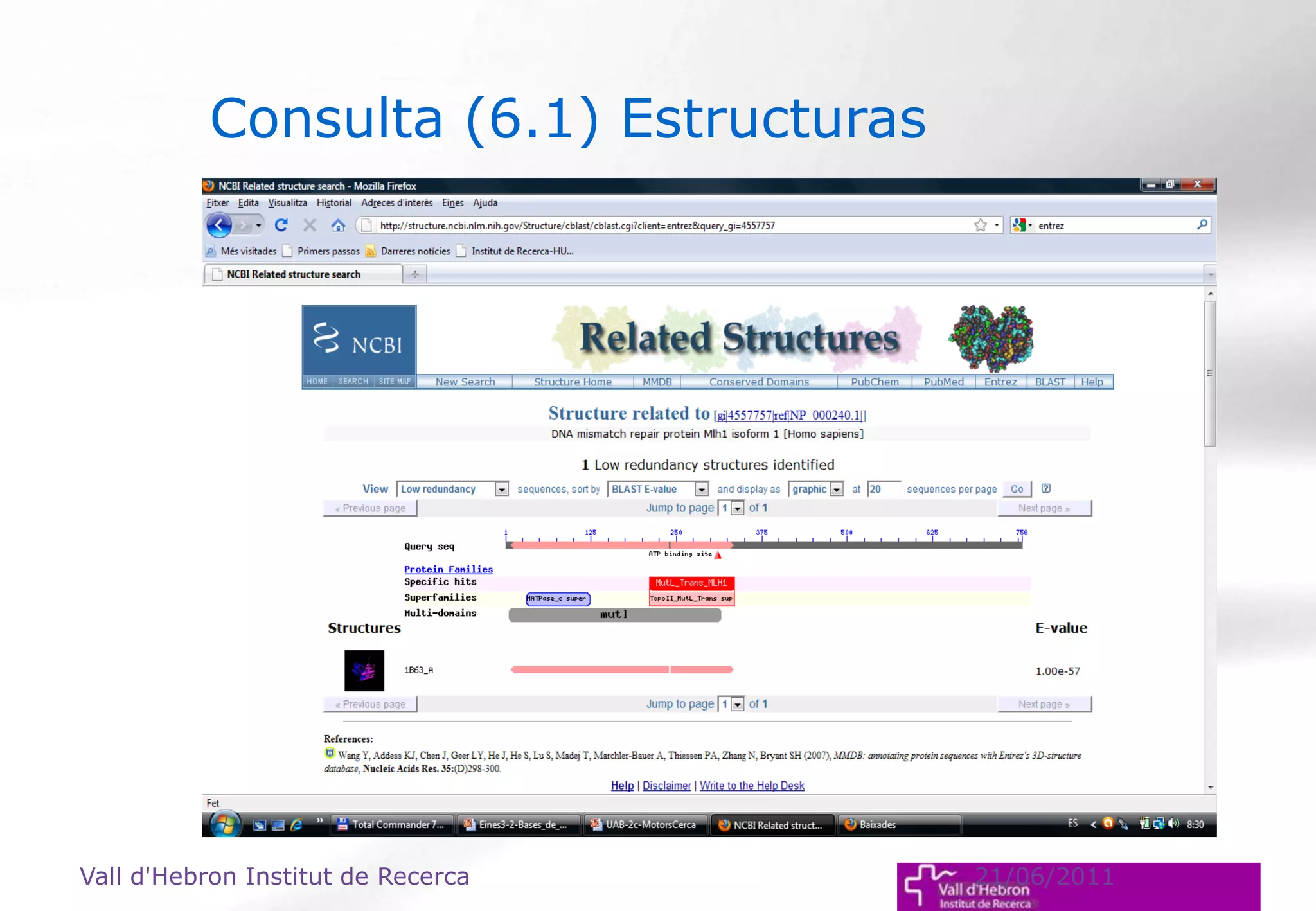
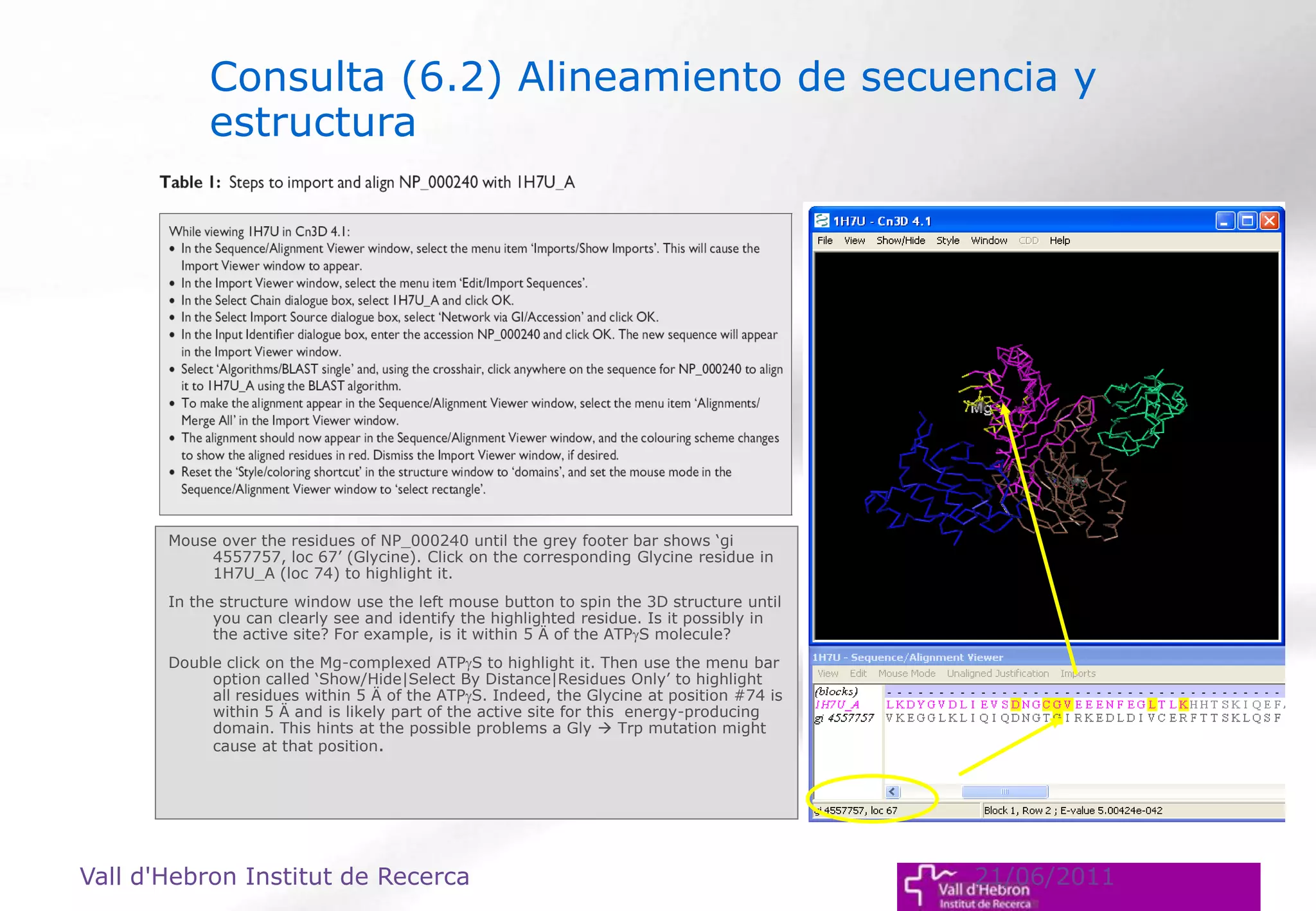
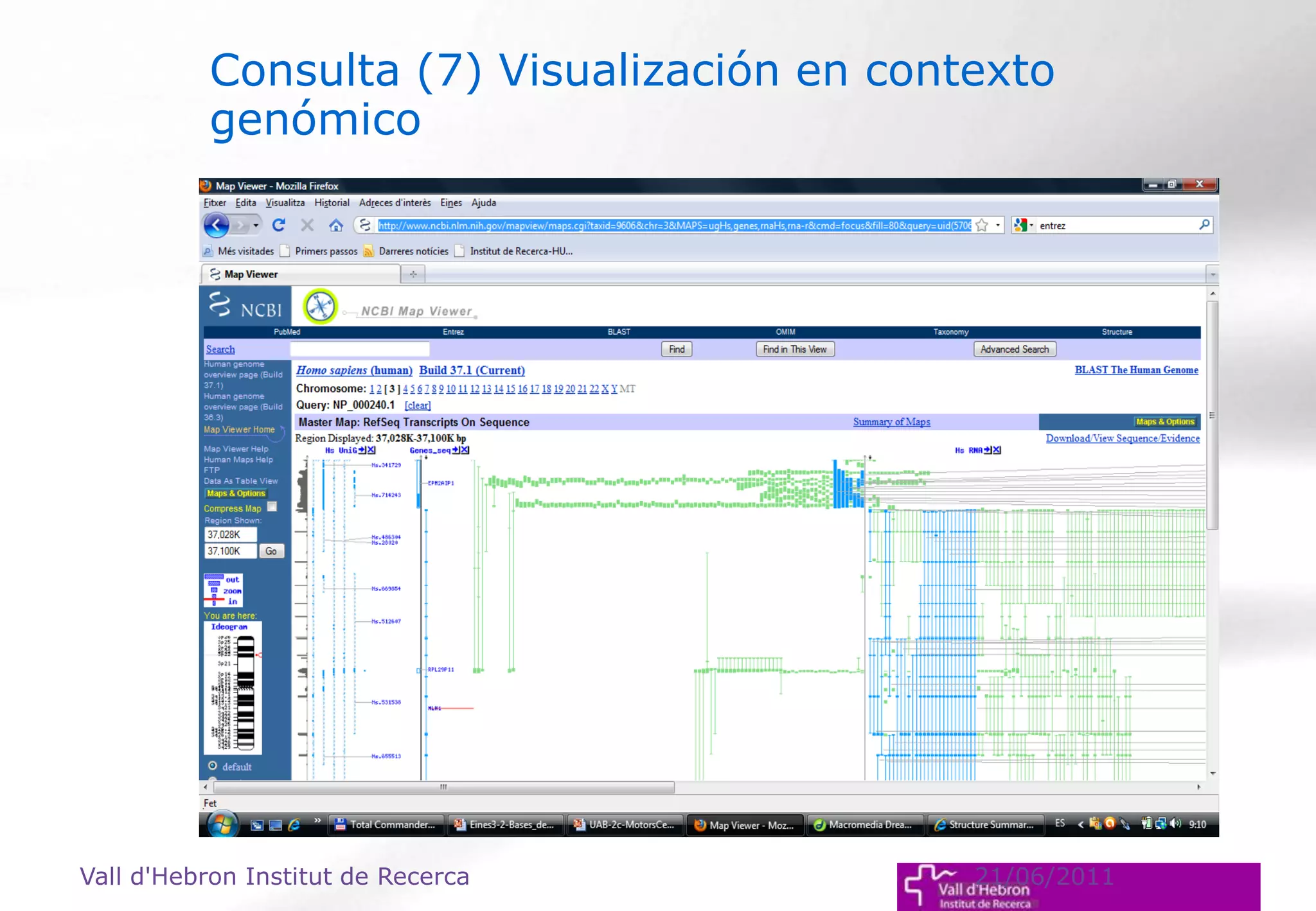
The document outlines the importance and structure of various biomedical databases relevant to genomics research, including types of databases, data submission processes, and tools for database exploitation. It details the roles of major data providers like NCBI and EBI, along with examples of nucleotide, genome, protein, and microarray databases. Additionally, it discusses the bioinformatics methods for data analysis and searching within these databases.