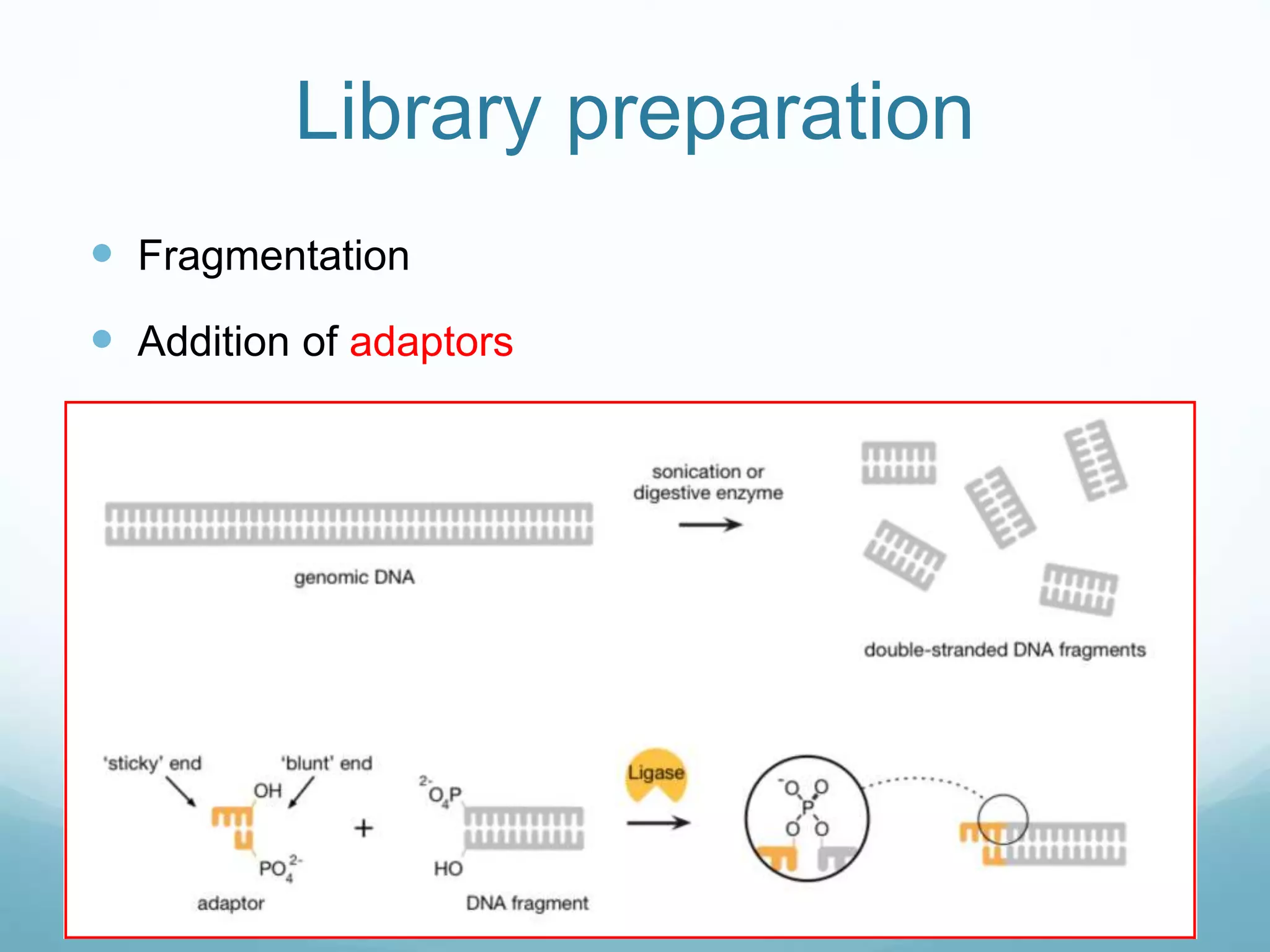
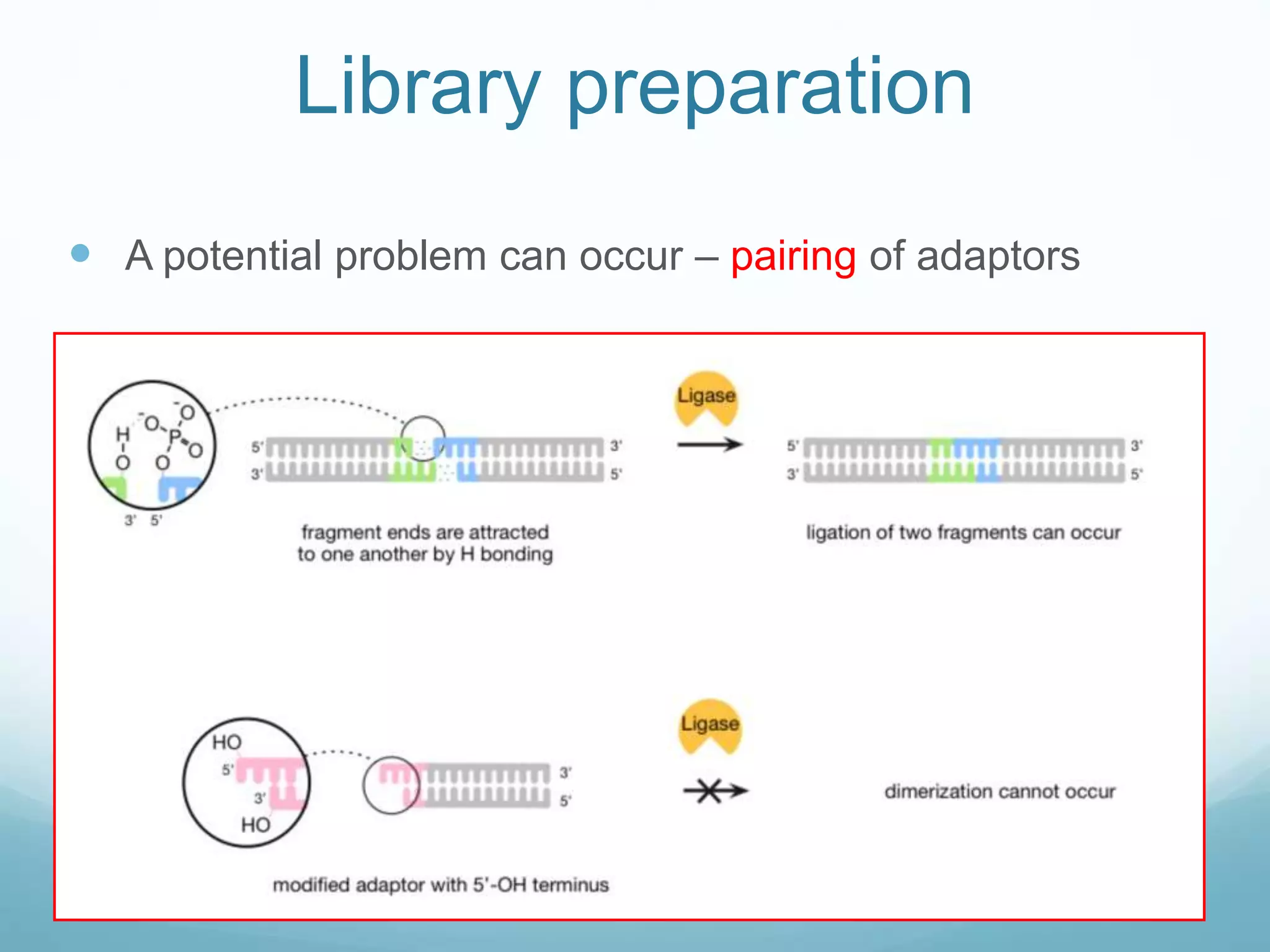
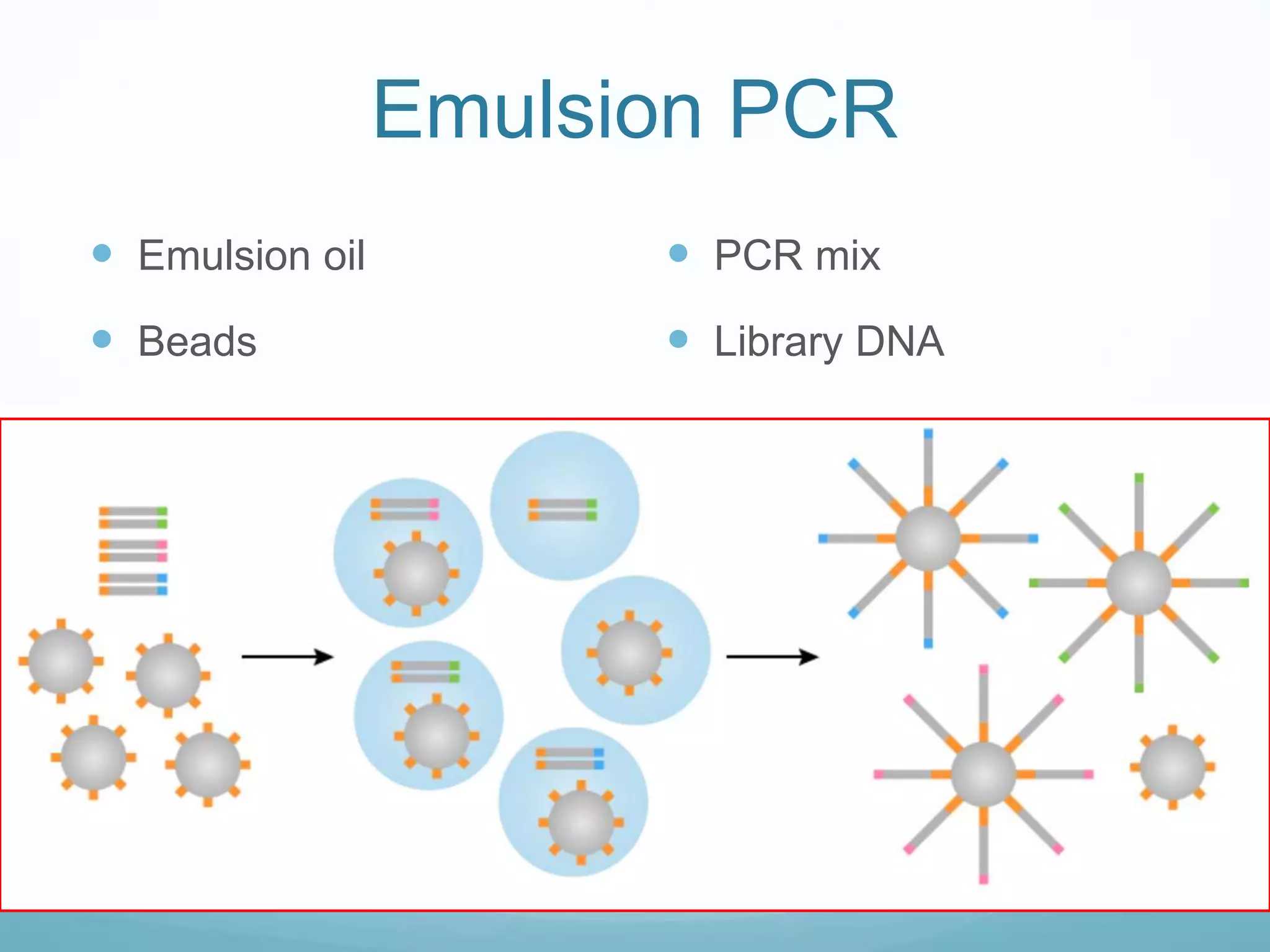
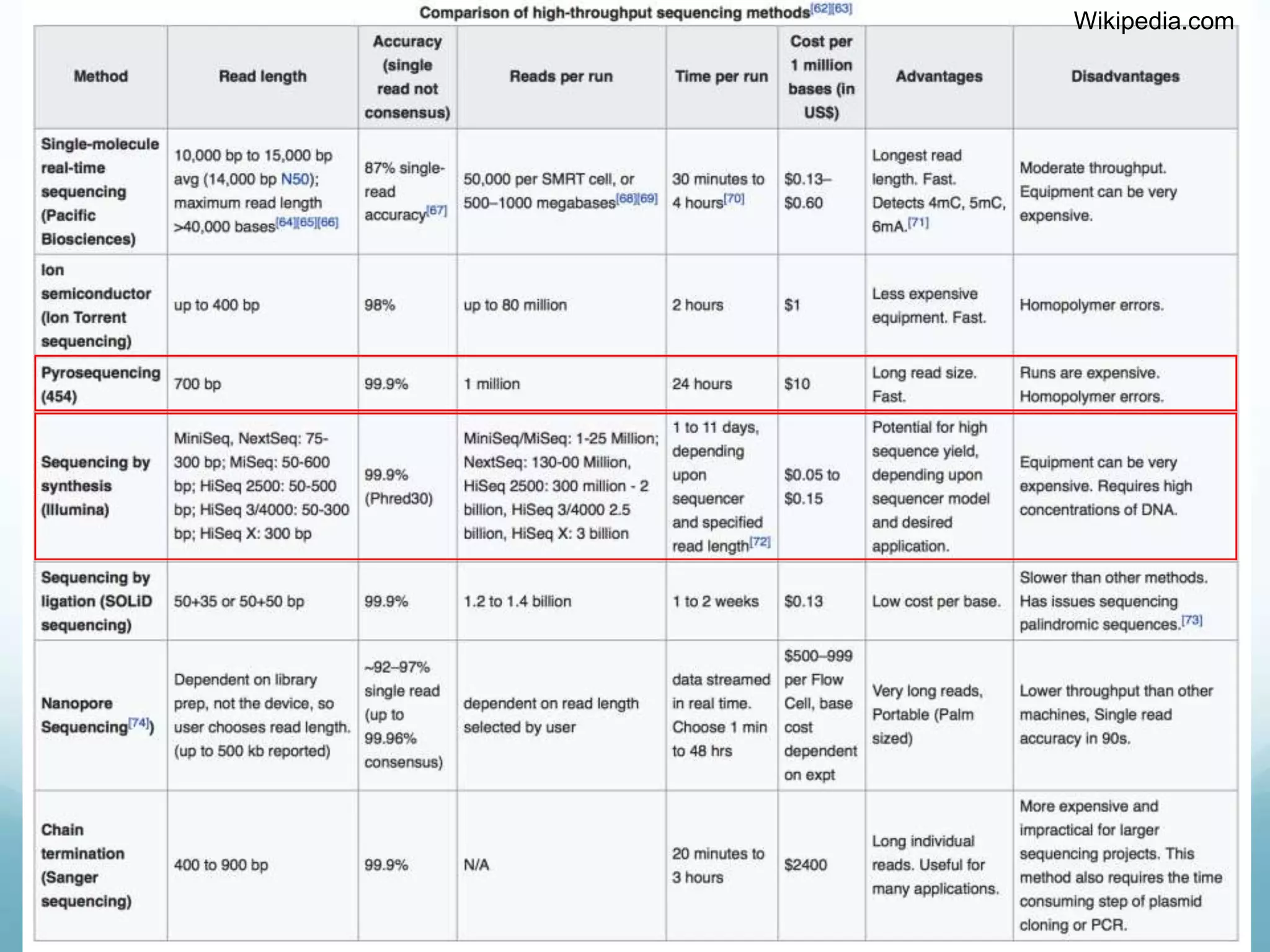
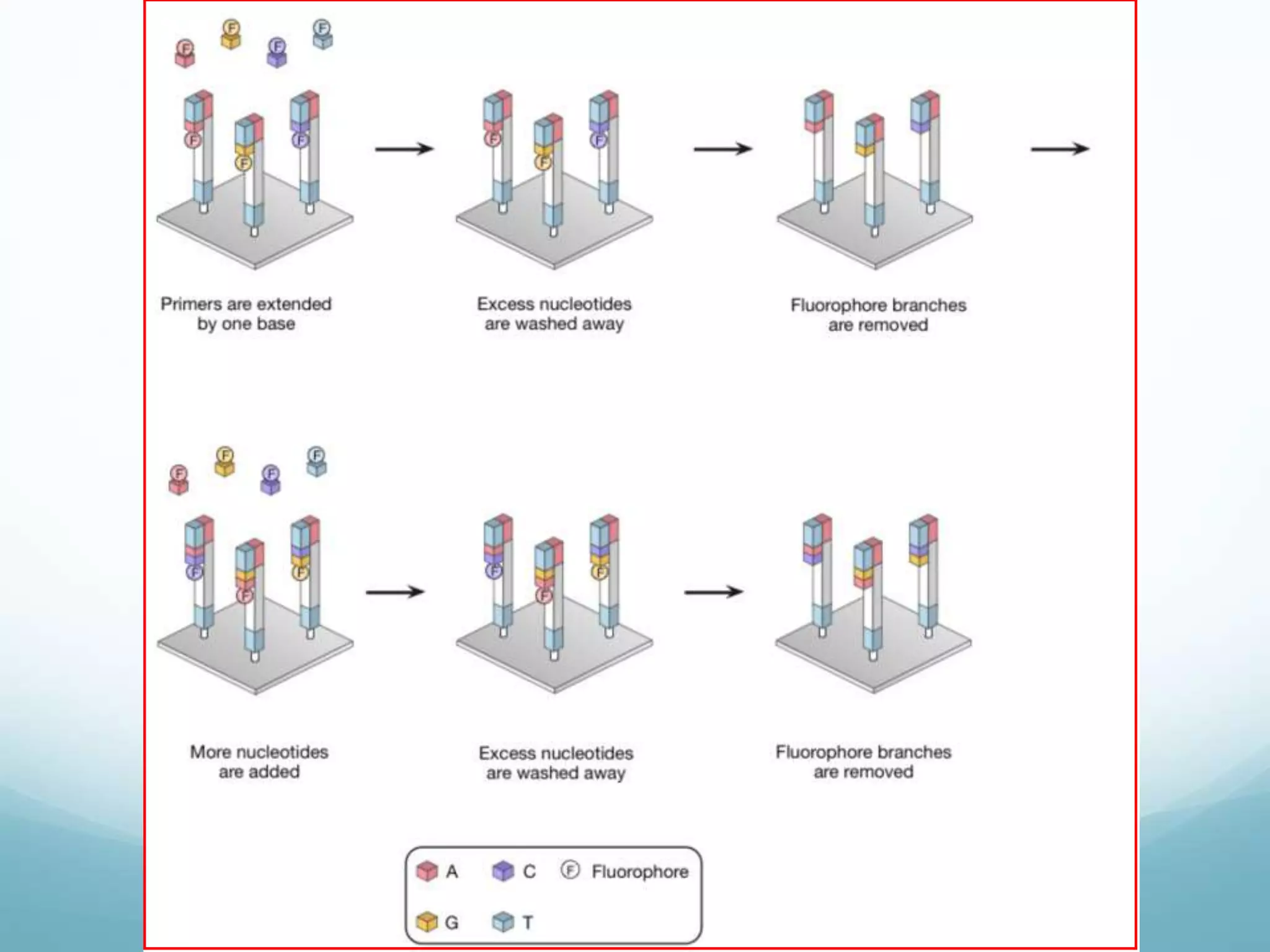
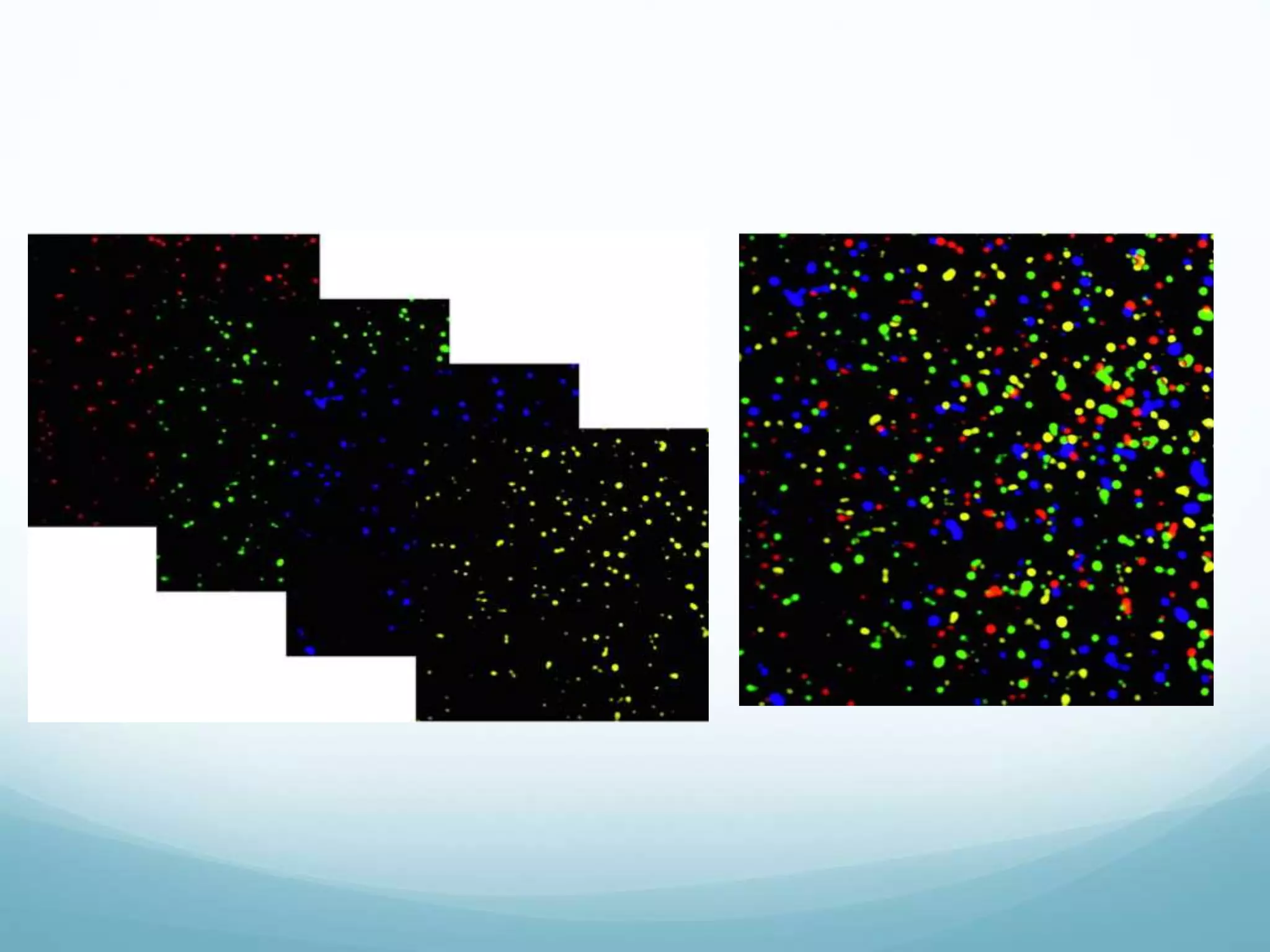
Next-generation sequencing (NGS) has revolutionized genomic research. NGS is faster, more accurate, and less expensive than traditional Sanger sequencing. The general steps for NGS involve library preparation through fragmentation and ligation of DNA with adapters, amplification, and sequencing. Several NGS platforms use reversible terminators to sequence DNA in a massively parallel fashion, identifying nucleotides based on detection of distinct fluorescent signals. Compared to first-generation sequencing, NGS allows for high-throughput, cost-effective sequencing of entire genomes.