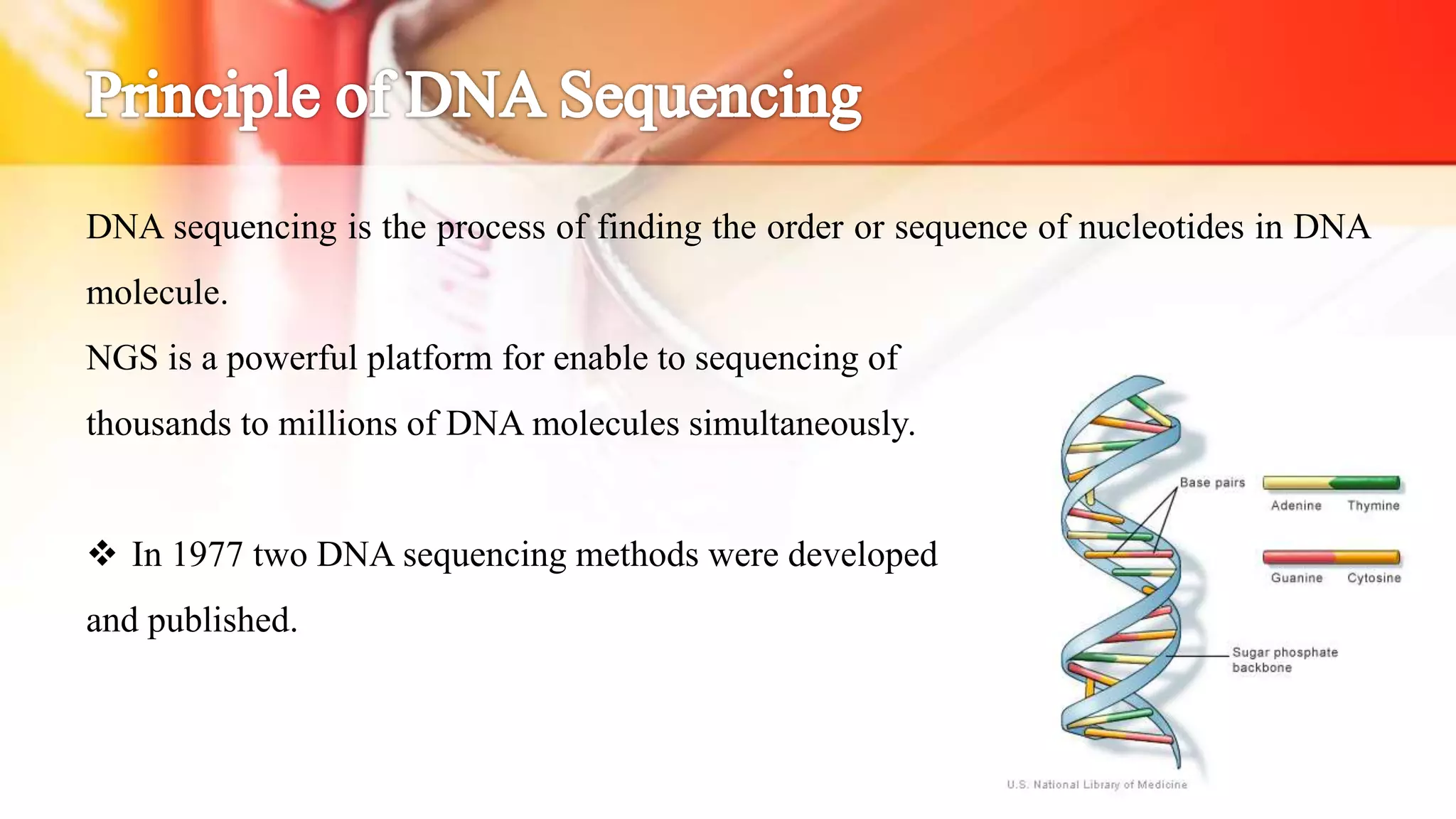
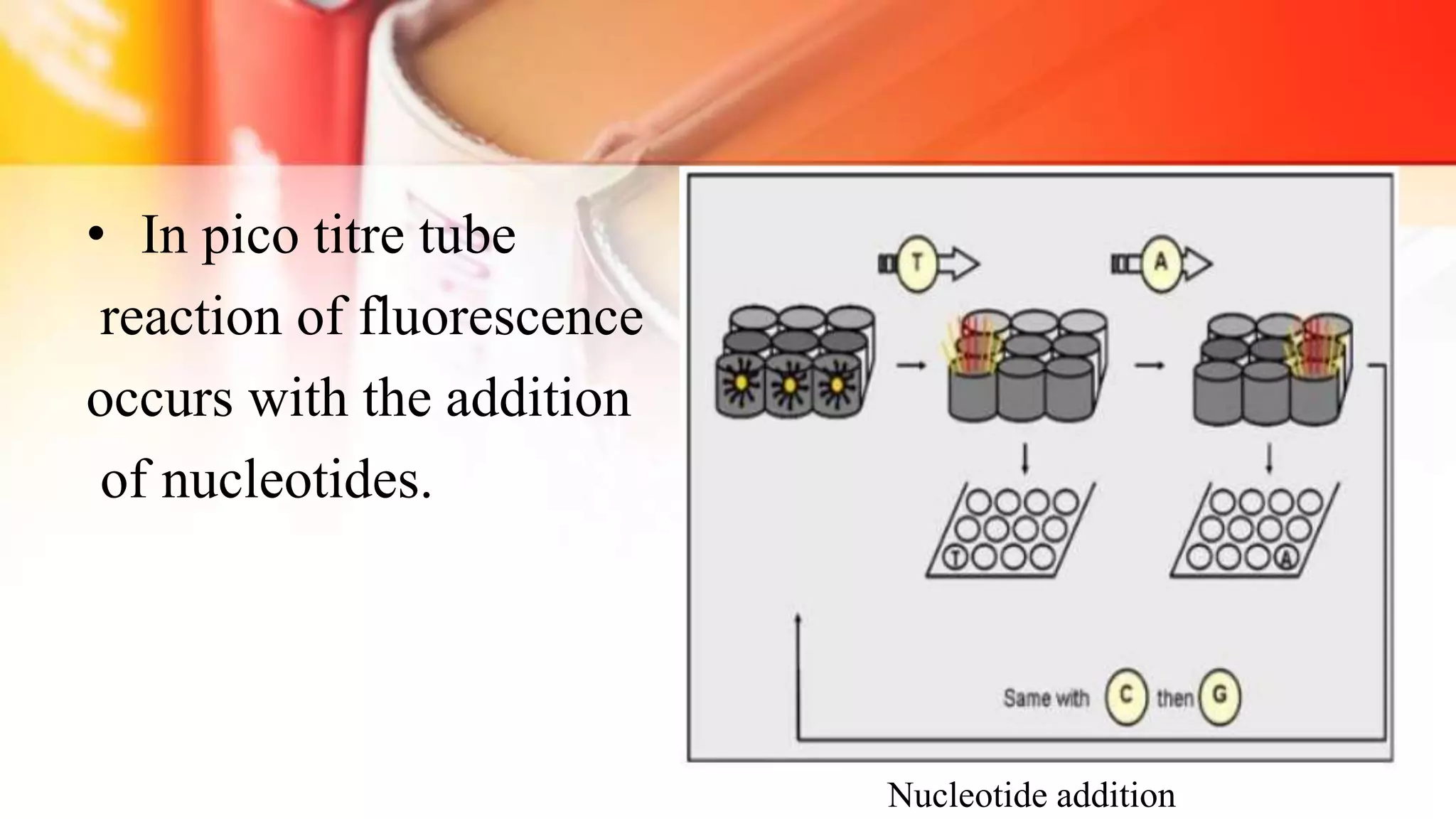
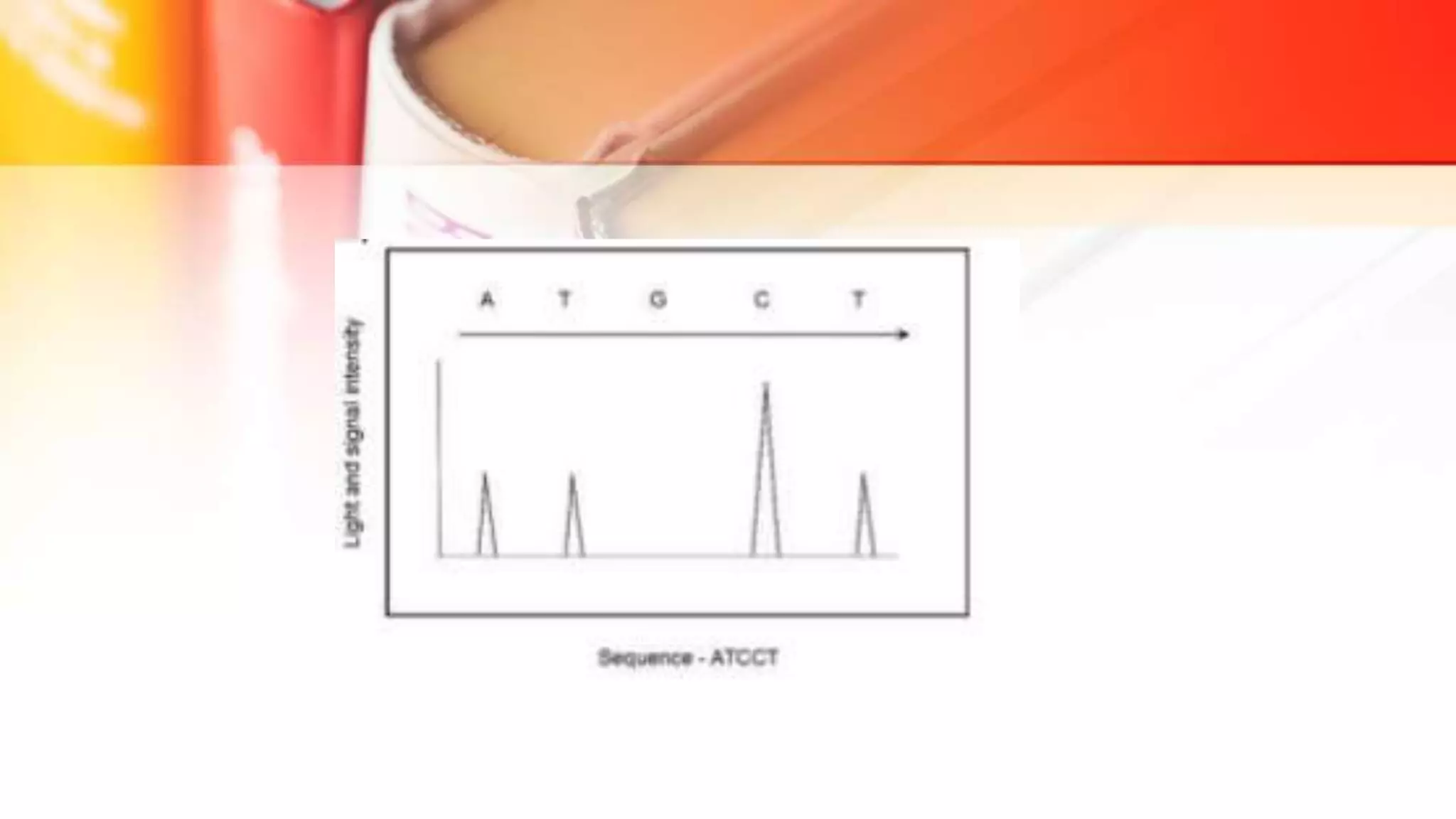
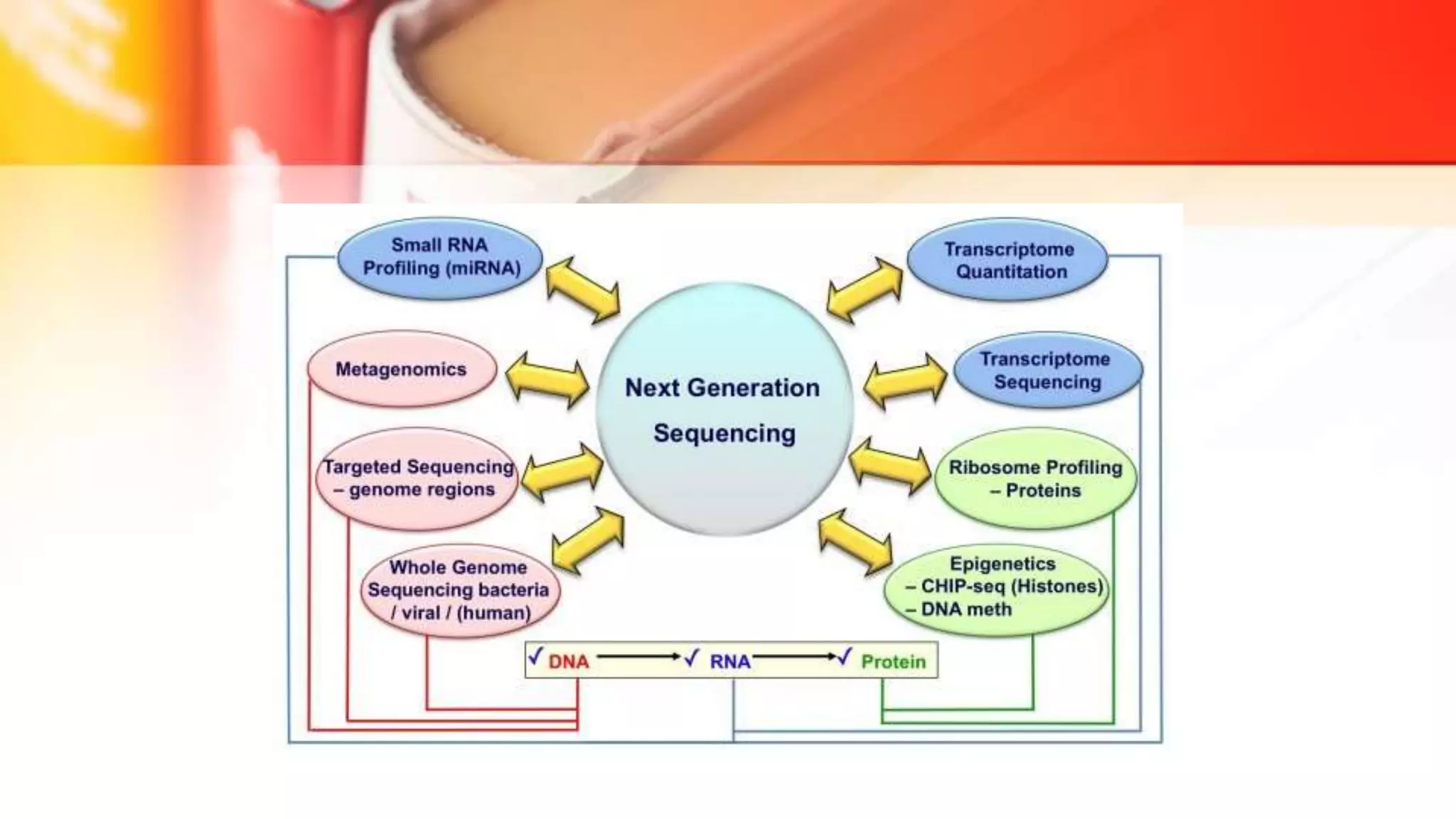
The document discusses the advancements in DNA sequencing, particularly focusing on Next Generation Sequencing (NGS) technologies, which allow for high-throughput and cost-effective sequencing of genomes. It outlines various sequencing methods, including Sanger and modern techniques like Illumina and Oxford Nanopore, emphasizing their roles in personalized medicine, genetic disease research, and cancer genomics. NGS has revolutionized molecular biology by enabling rapid sequencing and analysis of complex genetic information.