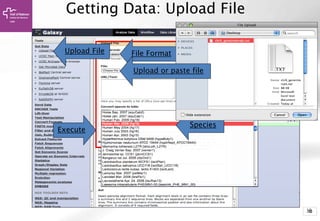
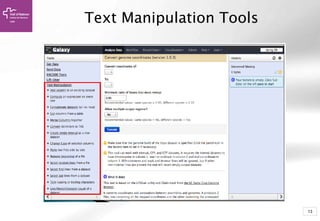
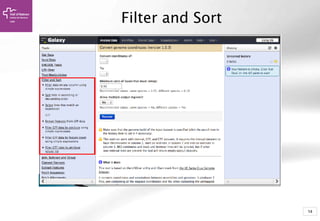
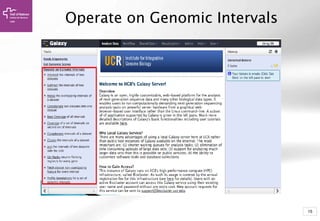
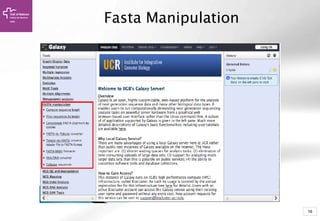
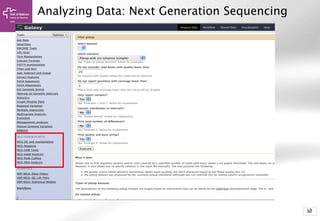
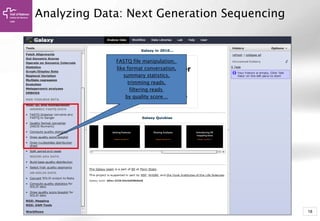
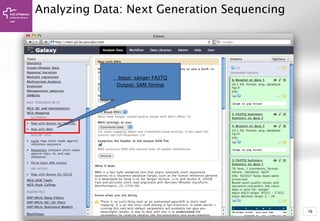
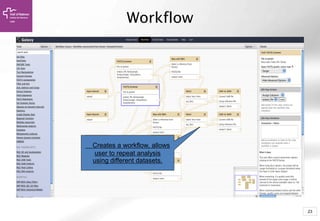
The document provides an introduction to Galaxy, a web-based platform for genome analysis that integrates various computational tools for bioinformatics. It covers the Galaxy interface, how to input and analyze data, and includes exercises for next-generation sequencing analysis. The document also outlines features like history tracking and workflow creation to facilitate repetitive analyses.






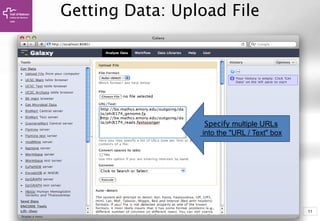
![99
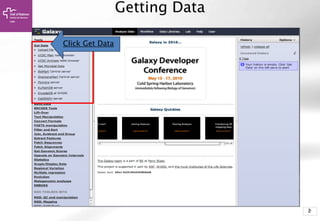
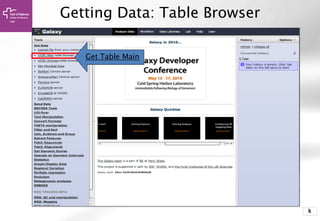
Getting Data: UCSC Table Browser
Get Output
clade: Mammal
genome: Human
assmbly: [current]
group: Genes and…
track: UCSC Genes
table: knownGene
region: position, chrX
Output format:
BED, and check
Send output to
Galaxy](https://image.slidesharecdn.com/bbr-2-140612013832-phpapp01/85/Introduction-to-Galaxy-UEB-UAT-Bioinformatics-Course-Session-2-2-VHIR-Barcelona-9-320.jpg)