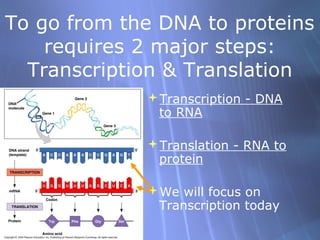
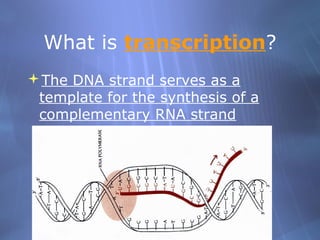
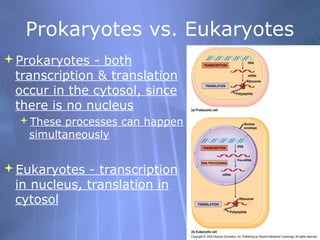
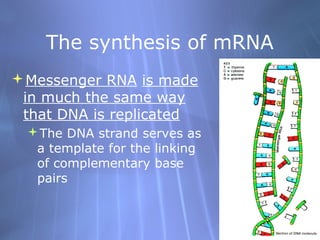
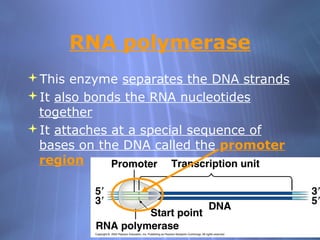
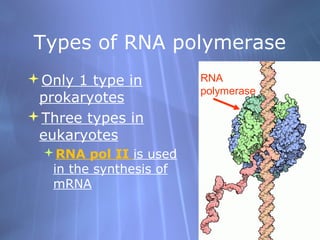
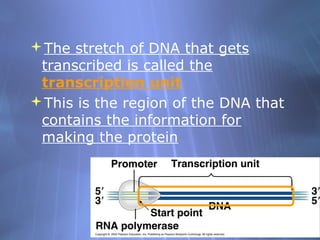
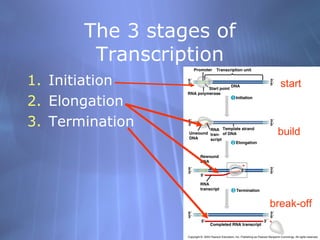
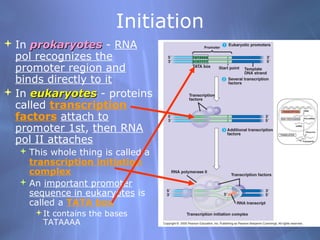
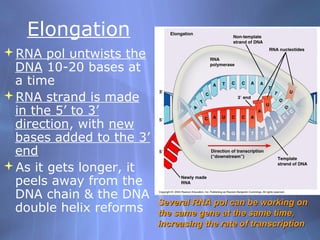
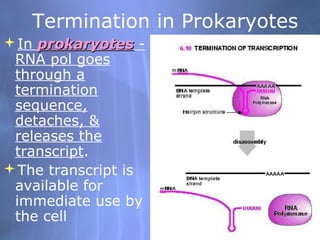
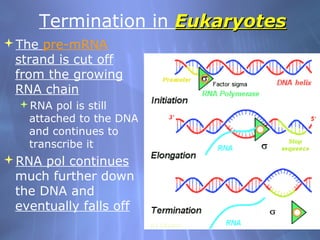
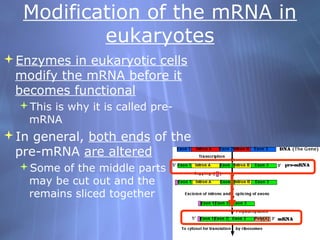
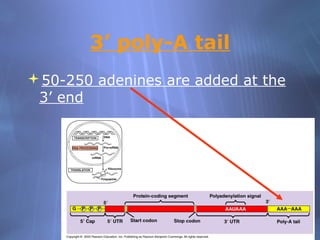
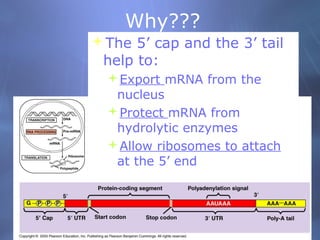
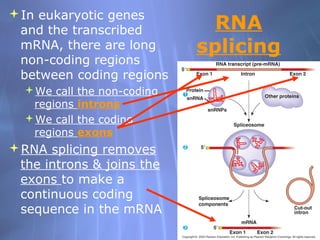
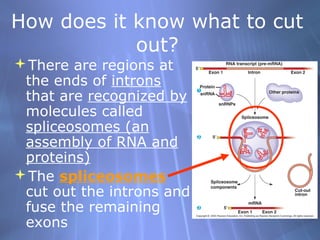
This document provides an overview of transcription in prokaryotes and eukaryotes. It discusses how DNA serves as a template for RNA production. The three main stages of transcription are initiation, elongation, and termination. In eukaryotes, the pre-mRNA undergoes processing including 5' capping, poly-A tail addition, and splicing of introns before becoming a functional mRNA that can be translated into a protein.