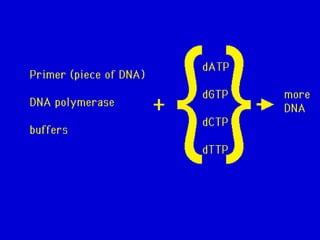
This document summarizes DNA replication. It describes the key requirements for replication like dNTPs, template DNA, primers, and magnesium ions. It explains that replication is semi-conservative, with each parental strand serving as a template for the daughter strands. It discusses the different models of replication and evidence from Meselson and Stahl's experiment supporting the semi-conservative model. It also provides details about replication proteins, the replication fork, and differences between prokaryotic and eukaryotic replication.