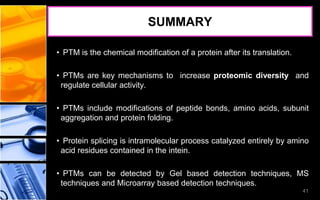
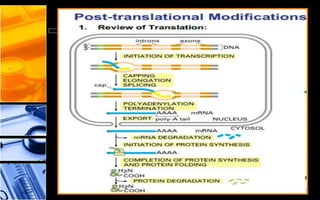
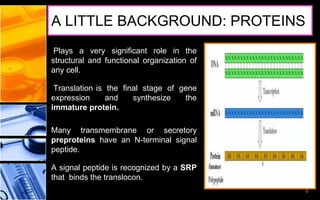
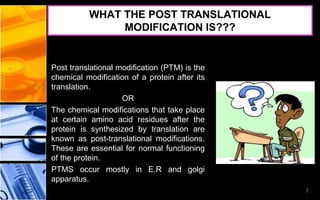
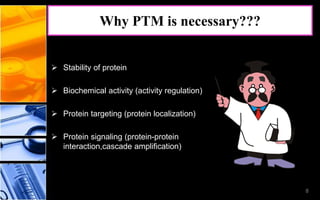
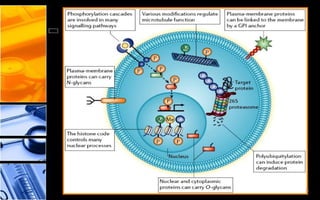
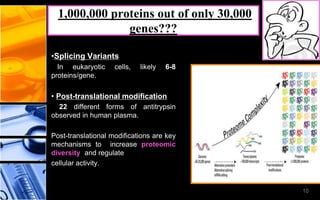
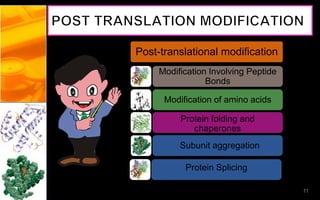
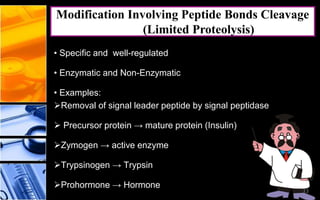
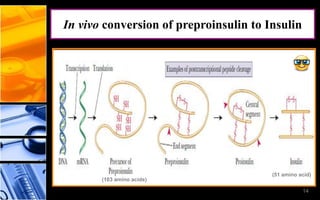
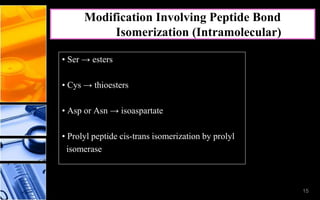
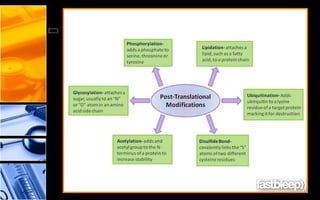
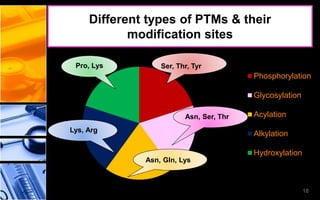
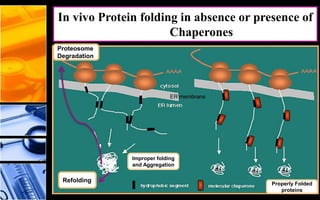
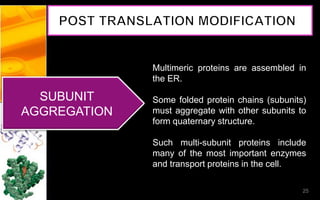
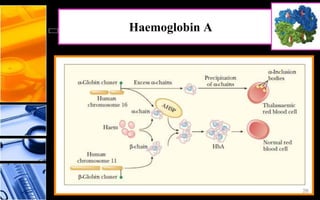
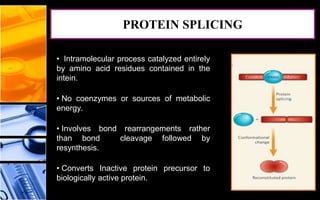
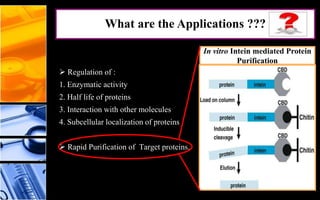
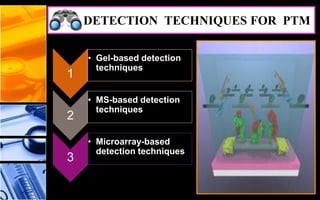
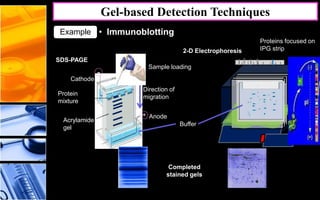
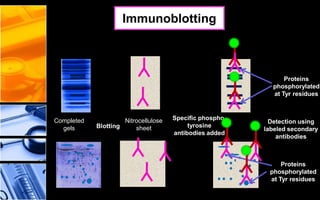
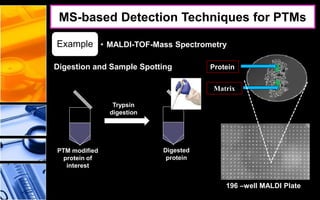
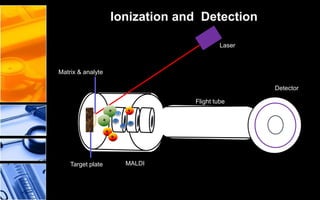
The document discusses post-translational modifications (PTMs), which are chemical modifications of proteins after translation, essential for their stability, activity, targeting, and signaling. It covers various types of PTMs, such as phosphorylation and glycosylation, their roles in protein diversity, and the mechanisms of protein splicing. Detection techniques for PTMs, including gel-based and mass spectrometry methods, are also outlined.






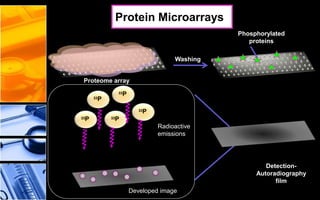
![Proteome array containing
potential substrates for
phosphorylation
Kinase
enzyme
[g-33P] ATP
solution
Protein
substrate
Kinase enzyme
[g-33P]
ATP
ADP
Ser
Phosphorylated
protein
Ser
Microarray-based Detection Techniques for
PTMs
• Protein MicroarraysExample](https://image.slidesharecdn.com/drefgjlruex1uhi6b1ru-140621003029-phpapp01/85/Post-translational-modifications-38-320.jpg)